1OKK
 
 | |
1OKO
 
 | | Crystal structure of Pseudomonas Aeruginosa Lectin 1 complexed with galactose at 1.6 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, PA-I GALACTOPHILIC LECTIN, ... | | Authors: | Cioci, G, Mitchell, E, Gautier, C, Wimmerova, M, Perez, S, Gilboa-Garber, N, Imberty, A. | | Deposit date: | 2003-07-28 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of Calcium and Galactose Recognition by the Lectin Pa-Il of Pseudomonas Aeruginosa
FEBS Lett., 555, 2003
|
|
1OKQ
 
 | | LAMININ ALPHA 2 CHAIN LG4-5 DOMAIN PAIR, CA1 SITE MUTANT | | Descriptor: | CALCIUM ION, LAMININ ALPHA 2 CHAIN | | Authors: | Wizemann, H, Garbe, J.H.O, Friedrich, M.V.K, Timpl, R, Sasaki, T, Hohenester, E. | | Deposit date: | 2003-07-28 | | Release date: | 2003-09-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Distinct Requirements for Heparin and Alpha-Dystroglycan Binding Revealed by Structure-Based Mutagenesis of the Laminin Alpha2 Lg4-Lg5 Domain Pair
J.Mol.Biol., 332, 2003
|
|
1OKS
 
 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
1OKT
 
 | | X-ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium falciparum | | Descriptor: | FORMIC ACID, GLUTATHIONE S-TRANSFERASE | | Authors: | Fritz-Wolf, K, Becker, A, Rahlfs, s, Harwaldt, P, Schirmer, R.H, Kabsch, W, Becker, K. | | Deposit date: | 2003-07-29 | | Release date: | 2003-11-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structure of Glutathione S-Transferase from the Malarial Parasite Plasmodium Falciparum
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1OKW
 
 | | Cyclin A binding groove inhibitor Ac-Arg-Arg-Leu-Asn-(m-Cl-Phe)-NH2 | | Descriptor: | ACE-ARG-ARG-LEU-ASN-FCL-NH2, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-07-31 | | Release date: | 2003-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OKX
 
 | | Binding Structure of Elastase Inhibitor Scyptolin A | | Descriptor: | ELASTASE 1, SCYPTOLIN A | | Authors: | Matern, U, Schleberger, C, Jelakovic, S, Weckesser, J, Schulz, G.E. | | Deposit date: | 2003-07-31 | | Release date: | 2003-10-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Binding Structure of Elastase Inhibitor Scyptolin A
Chem.Biol., 10, 2003
|
|
1OKY
 
 | | Structure of human PDK1 kinase domain in complex with staurosporine | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, GLYCEROL, STAUROSPORINE, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
1OKZ
 
 | | Structure of human PDK1 kinase domain in complex with UCN-01 | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE 1, 7-HYDROXYSTAUROSPORINE, GLYCEROL, ... | | Authors: | Komander, D, Kular, G.S, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2003-08-01 | | Release date: | 2004-07-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural Basis for Ucn-01 (7-Hydroxystaurosporine) Specificity and Pdk1 (3-Phosphoinositide-Dependent Protein Kinase-1) Inhibition
Biochem.J., 375, 2003
|
|
1OL0
 
 | | Crystal structure of a camelised human VH | | Descriptor: | GLYCEROL, IMMUNOGLOBULIN G, SULFATE ION | | Authors: | Dottorini, T, Vaughan, C.K, Walsh, M.A, Losurdo, P, Sollazzo, M. | | Deposit date: | 2003-08-02 | | Release date: | 2004-01-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of a Human Vh: Requirements for Maintaining a Monomeric Fragment
Biochemistry, 43, 2004
|
|
1OL1
 
 | | Cyclin A binding groove inhibitor H-Cit-Cit-Leu-Ile-(p-F-Phe)-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CIR-CIR-LEU-ILE-PFF-NH2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-08-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OL2
 
 | | Cyclin A binding groove inhibitor H-Arg-Arg-Leu-Asn-(p-F-Phe)-NH2 | | Descriptor: | ARG-ARG-LEU-ASN-PFF-NH2, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-08-05 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OL5
 
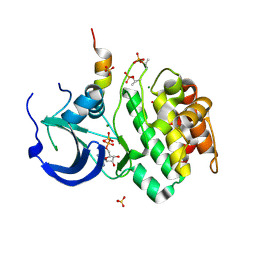 | | Structure of Aurora-A 122-403, phosphorylated on Thr287, Thr288 and bound to TPX2 1-43 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RESTRICTED EXPRESSION PROLIFERATION ASSOCIATED PROTEIN 100, ... | | Authors: | Bayliss, R, Conti, E. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Aurora-A Activation by Tpx2 at the Mitotic Spindle
Mol.Cell, 12, 2003
|
|
1OL6
 
 | |
1OL7
 
 | |
1OLC
 
 | |
1OLL
 
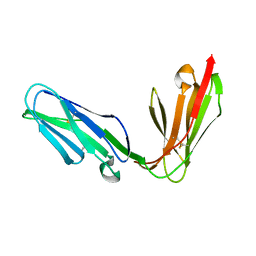 | | Extracellular region of the human receptor NKp46 | | Descriptor: | 1,2-ETHANEDIOL, NK RECEPTOR | | Authors: | Ponassi, M, Cantoni, C, Biassoni, R, Conte, R, Spallarossa, A, Pesce, A, Moretta, A, Moretta, L, Bolognesi, M, Bordo, D. | | Deposit date: | 2003-08-07 | | Release date: | 2003-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure of the Human Nk Cell Triggering Receptor Nkp46 Ectodomain
Biochem.Biophys.Res.Commun., 309, 2003
|
|
1OLO
 
 | |
1OLP
 
 | | Alpha Toxin from Clostridium Absonum | | Descriptor: | ALPHA-TOXIN, CALCIUM ION, ZINC ION | | Authors: | Briggs, D.C, Basak, A.K. | | Deposit date: | 2003-08-11 | | Release date: | 2003-10-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Clostridium Absonum Alpha-Toxin: New Insights Into Clostridial Phospholipase C Substrate Binding and Specificity
J.Mol.Biol., 333, 2003
|
|
1OLQ
 
 | | The Trichoderma reesei cel12a P201C mutant, structure at 1.7 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OLR
 
 | | The Humicola grisea Cel12A Enzyme Structure at 1.2 A Resolution | | Descriptor: | ENDO-BETA-1,4-GLUCANASE | | Authors: | Sandgren, M, Gualfetti, P.J, Shaw, A, Gross, L.S, Saldajeno, M, Berglund, G.I, Jones, T.A, Mitchinson, C. | | Deposit date: | 2003-08-11 | | Release date: | 2003-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The Humicola Grisea Cel12A Enzyme Structure at 1.2 A Resolution and the Impact of its Free Cysteine Residues on Thermal Stability
Protein Sci., 12, 2003
|
|
1OLS
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-12 | | Release date: | 2003-08-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1OLT
 
 | | Coproporphyrinogen III oxidase (HemN) from Escherichia coli is a Radical SAM enzyme. | | Descriptor: | IRON/SULFUR CLUSTER, OXYGEN-INDEPENDENT COPROPORPHYRINOGEN III OXIDASE, S-ADENOSYLMETHIONINE | | Authors: | Layer, G, Moser, J, Heinz, D.W, Jahn, D, Schubert, W.-D. | | Deposit date: | 2003-08-13 | | Release date: | 2003-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of Coproporphyrinogen III Oxidase Reveals Cofactor Geometry of Radical Sam Enzymes
Embo J., 22, 2003
|
|
1OLU
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-15 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
1OLX
 
 | | Roles of His291-alpha and His146-beta' in the reductive acylation reaction catalyzed by human branched-chain alpha-ketoacid dehydrogenase | | Descriptor: | 2-OXOISOVALERATE DEHYDROGENASE ALPHA SUBUNIT, 2-OXOISOVALERATE DEHYDROGENASE BETA SUBUNIT, GLYCEROL, ... | | Authors: | Wynn, R.M, Machius, M, Chuang, J.L, Li, J, Tomchick, D.R, Chuang, D.T. | | Deposit date: | 2003-08-18 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Roles of His291-Alpha and His146-Beta' in the Reductive Acylation Reaction Catalyzed by Human Branched-Chain Alpha-Ketoacid Dehydrogenase: Refined Phosphorylation Loop Structure in the Active Site.
J.Biol.Chem., 278, 2003
|
|
