1KNP
 
 | |
1KNQ
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KNR
 
 | | L-aspartate oxidase: R386L mutant | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, L-aspartate oxidase, ... | | Authors: | Bossi, R.T, Mattevi, A. | | Deposit date: | 2001-12-19 | | Release date: | 2002-04-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of FAD-bound L-aspartate oxidase: insight into substrate specificity and catalysis.
Biochemistry, 41, 2002
|
|
1KNU
 
 | | LIGAND BINDING DOMAIN OF THE HUMAN PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA IN COMPLEX WITH A SYNTHETIC AGONIST | | Descriptor: | (S)-3-(4-(2-CARBAZOL-9-YL-ETHOXY)-PHENYL)-2-ETHOXY-PROPIONIC ACID, PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA | | Authors: | Svensson, L.A, Mortensen, S.B, Fleckner, J, Woeldike, H.F. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Novel tricyclic-alpha-alkyloxyphenylpropionic acids: dual PPARalpha/gamma agonists with hypolipidemic and antidiabetic activity
J.MED.CHEM., 45, 2002
|
|
1KNV
 
 | | Bse634I restriction endonuclease | | Descriptor: | ACETATE ION, Bse634I restriction endonuclease, CHLORIDE ION | | Authors: | Grazulis, S, Deibert, M, Rimseliene, R, Skirgaila, R, Sasnauskas, G, Lagunavicius, A, Repin, V, Urbanke, C, Huber, R, Siksnys, V. | | Deposit date: | 2001-12-19 | | Release date: | 2002-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal structure of the Bse634I restriction endonuclease: comparison of two enzymes recognizing the same DNA sequence.
Nucleic Acids Res., 30, 2002
|
|
1KNX
 
 | | HPr kinase/phosphatase from Mycoplasma pneumoniae | | Descriptor: | Probable HPr(Ser) kinase/phosphatase | | Authors: | Allen, G.S. | | Deposit date: | 2001-12-19 | | Release date: | 2002-12-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of HPr Kinase/Phosphatase from Mycoplasma pneumoniae
J.Mol.Biol., 326, 2003
|
|
1KO1
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KO2
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with an oxidized Cys (cysteinesulfonic) | | Descriptor: | ACETATE ION, VIM-2 metallo-beta-lactamase, ZINC ION | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1KO3
 
 | | VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa with Cys221 reduced | | Descriptor: | ACETATE ION, CHLORIDE ION, HYDROXIDE ION, ... | | Authors: | Garcia-Saez, I, Docquier, J.-D, Rossolini, G.M, Dideberg, O. | | Deposit date: | 2001-12-20 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The three-dimensional structure of VIM-2, a Zn-beta-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form
J.Mol.Biol., 375, 2008
|
|
1KO4
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KO5
 
 | | Crystal structure of gluconate kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Gluconate kinase, MAGNESIUM ION | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KO8
 
 | | Crystal structure of gluconate kinase | | Descriptor: | 6-PHOSPHOGLUCONIC ACID, Gluconate kinase, MAGNESIUM ION | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOA
 
 | | TWITCHIN KINASE FRAGMENT (C.ELEGANS), AUTOREGULATED PROTEIN KINASE AND IMMUNOGLOBULIN DOMAINS | | Descriptor: | TWITCHIN | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOB
 
 | | TWITCHIN KINASE FRAGMENT (APLYSIA), AUTOREGULATED PROTEIN KINASE DOMAIN | | Descriptor: | TWITCHIN, VALINE | | Authors: | Kobe, B, Heierhorst, J, Feil, S.C, Parker, M.W, Benian, G.M, Weiss, K.R, Kemp, B.E. | | Deposit date: | 1996-06-28 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Giant protein kinases: domain interactions and structural basis of autoregulation.
EMBO J., 15, 1996
|
|
1KOE
 
 | | ENDOSTATIN | | Descriptor: | ENDOSTATIN | | Authors: | Hohenester, E, Sasaki, T, Olsen, B.R, Timpl, R. | | Deposit date: | 1998-01-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the angiogenesis inhibitor endostatin at 1.5 A resolution.
EMBO J., 17, 1998
|
|
1KOF
 
 | | Crystal structure of gluconate kinase | | Descriptor: | Gluconate kinase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
1KOG
 
 | | Crystal structure of E. coli threonyl-tRNA synthetase interacting with the essential domain of its mRNA operator | | Descriptor: | 5'-O-(N-(L-THREONYL)-SULFAMOYL)ADENOSINE, Threonyl-tRNA synthetase, Threonyl-tRNA synthetase mRNA, ... | | Authors: | Torres-Larrios, A, Dock-Bregeon, A.C, Romby, P, Rees, B, Sankaranarayanan, R, Caillet, J, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D. | | Deposit date: | 2001-12-20 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of translational control by Escherichia coli threonyl tRNA synthetase.
Nat.Struct.Biol., 9, 2002
|
|
1KOH
 
 | | THE CRYSTAL STRUCTURE AND MUTATIONAL ANALYSIS OF A NOVEL RNA-BINDING DOMAIN FOUND IN THE HUMAN TAP NUCLEAR MRNA EXPORT FACTOR | | Descriptor: | TIP ASSOCIATING PROTEIN | | Authors: | Ho, D.N, Coburn, G.A, Kang, Y, Cullen, B.R, Georgiadis, M.M. | | Deposit date: | 2001-12-20 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The crystal structure and mutational analysis of a novel RNA-binding domain found in the human Tap nuclear mRNA export factor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KOI
 
 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 FROM RHODNIUS PROLIXUS COMPLEXED WITH NITRIC OXIDE AT 1.08 A RESOLUTION | | Descriptor: | NITRIC OXIDE, NITROPHORIN 4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roberts, S.A, Weichsel, A, Qiu, Y, Shelnutt, J.A, Walker, F.A, Montfort, W.R. | | Deposit date: | 2001-05-03 | | Release date: | 2002-01-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ligand-induced heme ruffling and bent no geometry in ultra-high-resolution structures of nitrophorin 4.
Biochemistry, 40, 2001
|
|
1KOJ
 
 | | Crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonohydroxamic acid | | Descriptor: | 5-PHOSPHO-D-ARABINOHYDROXAMIC ACID, Glucose-6-phosphate isomerase | | Authors: | Arsenieva, D, Hardre, R, Salmon, L, Jeffery, C.J. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of rabbit phosphoglucose isomerase complexed with 5-phospho-D-arabinonohydroxamic acid.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KOO
 
 | | THE CRYSTAL STRUCTURE AND MUTATIONAL ANALYSIS OF A NOVEL RNA-BINDING DOMAIN FOUND IN THE HUMAN TAP NUCLEAR MRNA EXPORT FACTOR | | Descriptor: | TIP ASSOCIATING PROTEIN | | Authors: | Ho, D.N, Coburn, G.A, Kang, Y, Cullen, B.R, Georgiadis, M.M. | | Deposit date: | 2001-12-21 | | Release date: | 2002-02-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The crystal structure and mutational analysis of a novel RNA-binding domain found in the human Tap nuclear mRNA export factor.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1KOR
 
 | | Crystal Structure of Thermus thermophilus HB8 Argininosuccinate Synthetase in complex with inhibitors | | Descriptor: | ARGININE, Argininosuccinate Synthetase, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Goto, M, Nakajima, Y, Hirotsu, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of argininosuccinate synthetase from Thermus thermophilus HB8. Structural basis for the catalytic action.
J.Biol.Chem., 277, 2002
|
|
1KOU
 
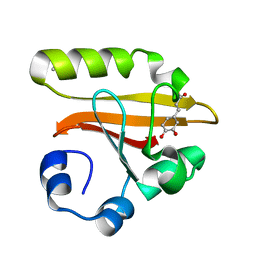 | | Crystal Structure of the Photoactive Yellow Protein Reconstituted with Caffeic Acid at 1.16 A Resolution | | Descriptor: | CAFFEIC ACID, N-BUTANE, PHOTOACTIVE YELLOW PROTEIN | | Authors: | van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2001-12-22 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structure of the photoactive yellow protein reconstituted with caffeic acid at 1.16 A resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KP0
 
 | |
1KP2
 
 | |
