1DGR
 
 | |
1DGS
 
 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
1DGW
 
 | |
1DHS
 
 | |
1DI2
 
 | |
1DI8
 
 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE | | Descriptor: | 4-[3-HYDROXYANILINO]-6,7-DIMETHOXYQUINAZOLINE, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Shewchuk, L, Hassell, A, Kuyper, L.F. | | Deposit date: | 1999-11-29 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase.
J.Med.Chem., 43, 2000
|
|
1DI9
 
 | | THE STRUCTURE OF P38 MITOGEN-ACTIVATED PROTEIN KINASE IN COMPLEX WITH 4-[3-METHYLSULFANYLANILINO]-6,7-DIMETHOXYQUINAZOLINE | | Descriptor: | 4-[3-METHYLSULFANYLANILINO]-6,7-DIMETHOXYQUINAZOLINE, P38 KINASE | | Authors: | Shewchuk, L, Hassell, A, Kuyper, L.F. | | Deposit date: | 1999-11-29 | | Release date: | 2000-11-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding mode of the 4-anilinoquinazoline class of protein kinase inhibitor: X-ray crystallographic studies of 4-anilinoquinazolines bound to cyclin-dependent kinase 2 and p38 kinase.
J.Med.Chem., 43, 2000
|
|
1DIA
 
 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY249543 | | Descriptor: | METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [[[2-AMINO-5,6,7,8-TETRAHYDRO-4-HYDROXY-PYRIDO[2,3-D]PYRIMIDIN-6-YL]-ETHYL]-PHENYL]-CARBONYL-GLUTAMIC ACID | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIB
 
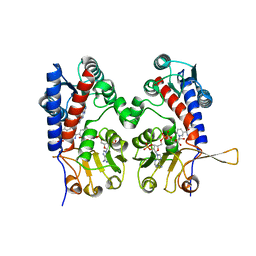 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY345899 | | Descriptor: | 4-(7-AMINO-9-HYDROXY-1-OXO-3,3A,4,5-TETRAHYDRO-2,5,6,8,9B-PENTAAZA-CYCLOPENTA[A]NAPHTHALEN-2-YL)-PHENYLCARBONYL-GLUTAMI C ACID, METHYLENETETRAHYDROFOLATE DEHYDROGENASE/CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DIG
 
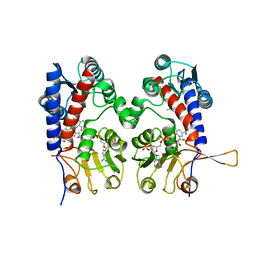 | | HUMAN METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE COMPLEXED WITH NADP AND INHIBITOR LY374571 | | Descriptor: | ACETATE ION, METHYLENETETRAHYDROFOLATE DEHYDROGENASE / CYCLOHYDROLASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schmidt, A, Wu, H, MacKenzie, R.E, Chen, V.J, Bewly, J.R, Ray, J.E, Toth, J.E, Cygler, M. | | Deposit date: | 1999-11-29 | | Release date: | 2000-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase.
Biochemistry, 39, 2000
|
|
1DII
 
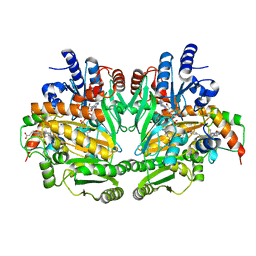 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE AT 2.5 A RESOLUTION | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.N, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DIO
 
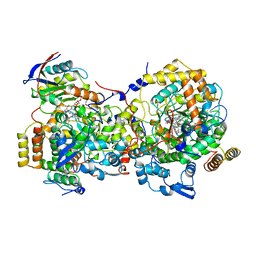 | | DIOL DEHYDRATASE-CYANOCOBALAMIN COMPLEX FROM KLEBSIELLA OXYTOCA | | Descriptor: | COBALAMIN, POTASSIUM ION, PROTEIN (DIOL DEHYDRATASE), ... | | Authors: | Shibata, N, Masuda, J, Tobimatsu, T, Toraya, T, Suto, K, Morimoto, Y, Yasuoka, N. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase.
Structure Fold.Des., 7, 1999
|
|
1DIQ
 
 | | CRYSTAL STRUCTURE OF P-CRESOL METHYLHYDROXYLASE WITH SUBSTRATE BOUND | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, HEME C, ... | | Authors: | Cunane, L.M, Chen, Z.W, Shamala, N, Mathews, F.S, Cronin, C.S, McIntire, W.S. | | Deposit date: | 1999-11-29 | | Release date: | 1999-12-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structures of the flavocytochrome p-cresol methylhydroxylase and its enzyme-substrate complex: gated substrate entry and proton relays support the proposed catalytic mechanism.
J.Mol.Biol., 295, 2000
|
|
1DIR
 
 | | CRYSTAL STRUCTURE OF A MONOCLINIC FORM OF DIHYDROPTERIDINE REDUCTASE FROM RAT LIVER | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Su, Y, Skinner, M.M, Matthews, D.A, Whitely, J.M, Xuong, N.H. | | Deposit date: | 1994-04-18 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a monoclinic form of dihydropteridine reductase from rat liver.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1DIY
 
 | | CRYSTAL STRUCTURE OF ARACHIDONIC ACID BOUND IN THE CYCLOOXYGENASE ACTIVE SITE OF PGHS-1 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, ... | | Authors: | Malkowski, M.G, Ginell, S.L, Smith, W.L, Garavito, R.M. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The productive conformation of arachidonic acid bound to prostaglandin synthase.
Science, 289, 2000
|
|
1DJ6
 
 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
1DJ7
 
 | | CRYSTAL STRUCTURE OF FERREDOXIN THIOREDOXIN REDUCTASE | | Descriptor: | FERREDOXIN THIOREDOXIN REDUCTASE: CATALYTIC CHAIN, FERREDOXIN THIOREDOXIN REDUCTASE: VARIABLE CHAIN, IRON/SULFUR CLUSTER, ... | | Authors: | Dai, S, Schwendtmayer, C, Schurmann, P, Ramaswamy, S, Eklund, H. | | Deposit date: | 1999-12-02 | | Release date: | 2000-02-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox signaling in chloroplasts: cleavage of disulfides by an iron-sulfur cluster.
Science, 287, 2000
|
|
1DJ8
 
 | |
1DJH
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH BARIUM | | Descriptor: | ACETATE ION, BARIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1DJI
 
 | | PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C-DELTA1 FROM RAT COMPLEXED WITH CALCIUM | | Descriptor: | ACETATE ION, CALCIUM ION, PHOSPHOINOSITIDE-SPECIFIC PHOSPHOLIPASE C, ... | | Authors: | Essen, L.-O, Perisic, O, Williams, R.L. | | Deposit date: | 1996-09-25 | | Release date: | 1997-07-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-delta1.
Biochemistry, 36, 1997
|
|
1DJN
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT WILD TYPE TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJQ
 
 | | STRUCTURAL AND BIOCHEMICAL CHARACTERIZATION OF RECOMBINANT C30A MUTANT OF TRIMETHYLAMINE DEHYDROGENASE FROM METHYLOPHILUS METHYLOTROPHUS (SP. W3A1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, IRON/SULFUR CLUSTER, ... | | Authors: | Trickey, P, Basran, J, Lian, L.-Y, Chen, Z.-W, Barton, J.D, Sutcliffe, M.J, Scrutton, N.S, Mathews, F.S. | | Deposit date: | 1999-12-03 | | Release date: | 1999-12-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and biochemical characterization of recombinant wild type and a C30A mutant of trimethylamine dehydrogenase from methylophilus methylotrophus (sp. W(3)A(1)).
Biochemistry, 39, 2000
|
|
1DJR
 
 | | HEAT-LABILE ENTEROTOXIN B-PENTAMER COMPLEXED WITH M-CARBOXYPHENYL-ALPHA-D-GALACTOSE | | Descriptor: | BENZOIC ACID, GLYCEROL, HEAT-LABILE ENTEROTOXIN, ... | | Authors: | Minke, W.E, Pickens, J, Merritt, E.A, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 1999-12-03 | | Release date: | 2000-06-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of m-carboxyphenyl-alpha-D-galactopyranoside complexed to heat-labile enterotoxin at 1.3 A resolution: surprising variations in ligand-binding modes.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1DJT
 
 | |
1DJU
 
 | | CRYSTAL STRUCTURE OF AROMATIC AMINOTRANSFERASE FROM PYROCOCCUS HORIKOSHII OT3 | | Descriptor: | AROMATIC AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matsui, I, Matsui, E, Sakai, Y, Kikuchi, H, Kawarabayashi, H. | | Deposit date: | 1999-12-06 | | Release date: | 2001-04-11 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular structure of hyperthermostable aromatic aminotransferase with novel substrate specificity from Pyrococcus horikoshii.
J.Biol.Chem., 275, 2000
|
|
