1C41
 
 | | CRYSTAL STRUCTURES OF A PENTAMERIC FUNGAL AND AN ICOSAHEDRAL PLANT LUMAZINE SYNTHASE REVEALS THE STRUCTURAL BASIS FOR DIFFERENCES IN ASSEMBLY | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, LUMAZINE SYNTHASE, SULFATE ION | | Authors: | Persson, K, Schneider, G, Jordan, D.B, Viitanen, P.V, Sandalova, T. | | Deposit date: | 1999-08-03 | | Release date: | 2000-08-06 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure analysis of a pentameric fungal and an icosahedral plant lumazine synthase reveals the structural basis for differences in assembly
Protein Sci., 8, 1999
|
|
1C44
 
 | |
1C47
 
 | |
1C48
 
 | | MUTATED SHIGA-LIKE TOXIN B SUBUNIT (G62T) | | Descriptor: | PROTEIN (SHIGA-LIKE TOXIN I B SUBUNIT) | | Authors: | Ling, H, Bast, D, Brunton, J.L, Read, R.J. | | Deposit date: | 1999-08-11 | | Release date: | 2000-08-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of the Primary Receptor Binding Site of Shiga-Like Toxin B Subunits: Structures of Mutated Shiga-Like Toxin I B-Pentamer with and without Bound Carbohydrate
To be Published
|
|
1C4F
 
 | | GREEN FLUORESCENT PROTEIN S65T AT PH 4.6 | | Descriptor: | GREEN FLUORESCENT PROTEIN | | Authors: | Elsliger, M.A, Wachter, R.M, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1999-08-21 | | Release date: | 1999-08-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and spectral response of green fluorescent protein variants to changes in pH.
Biochemistry, 38, 1999
|
|
1C4G
 
 | |
1C4K
 
 | | ORNITHINE DECARBOXYLASE MUTANT (GLY121TYR) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, PROTEIN (ORNITHINE DECARBOXYLASE), PYRIDOXAL-5'-PHOSPHATE | | Authors: | Vitali, J, Hackert, M.L. | | Deposit date: | 1999-08-26 | | Release date: | 2000-02-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Three-dimensional structure of the Gly121Tyr dimeric form of ornithine decarboxylase from Lactobacillus 30a.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1C4Q
 
 | |
1C4W
 
 | | 1.9 A STRUCTURE OF A-THIOPHOSPHONATE MODIFIED CHEY D57C | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Halkides, C.J, McEvoy, M.M, Matsumura, P, Volz, K, Dahlquist, F.W. | | Deposit date: | 1999-09-28 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The 1.9 A resolution crystal structure of phosphono-CheY, an analogue of the active form of the response regulator, CheY.
Biochemistry, 39, 2000
|
|
1C57
 
 | | DIRECT DETERMINATION OF THE POSITIONS OF DEUTERIUM ATOMS OF BOUND WATER IN CONCANAVALIN A BY NEUTRON LAUE CRYSTALLOGRAPHY | | Descriptor: | CALCIUM ION, Concanavalin-Br, MANGANESE (II) ION | | Authors: | Habash, J, Raftery, J, Nuttall, R, Price, H.J, Lehmann, M.S, Wilkinson, C, Kalb, A.J, Helliwell, J.R. | | Deposit date: | 1999-10-26 | | Release date: | 2000-05-08 | | Last modified: | 2023-12-27 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Direct determination of the positions of the deuterium atoms of the bound water in -concanavalin A by neutron Laue crystallography.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C5B
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8 UNLIGANDED FORM | | Descriptor: | CHIMERIC DECARBOXYLASE ANTIBODY 21D8 | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-08 | | Release date: | 2000-10-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5C
 
 | | DECARBOXYLASE CATALYTIC ANTIBODY 21D8-HAPTEN COMPLEX | | Descriptor: | 2-ACETYLAMINO-NAPTHALENE-1,5-DISULFONIC ACID, CHIMERIC DECARBOXYLASE ANTIBODY 21D8, GLYCEROL | | Authors: | Hotta, K, Wilson, I.A. | | Deposit date: | 1999-11-09 | | Release date: | 2000-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Catalysis of decarboxylation by a preorganized heterogeneous microenvironment: crystal structures of abzyme 21D8.
J.Mol.Biol., 302, 2000
|
|
1C5E
 
 | | BACTERIOPHAGE LAMBDA HEAD PROTEIN D | | Descriptor: | GLYCEROL, HEAD DECORATION PROTEIN | | Authors: | Yang, F, Forrer, P, Dauter, Z, Pluckthun, A, Wlodawer, A. | | Deposit date: | 1999-11-18 | | Release date: | 2000-03-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD.
Nat.Struct.Biol., 7, 2000
|
|
1C5F
 
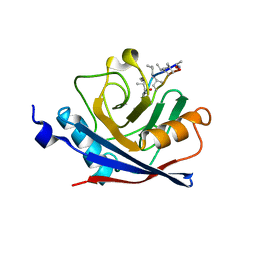 | | CRYSTAL STRUCTURE OF THE CYCLOPHILIN-LIKE DOMAIN FROM BRUGIA MALAYI COMPLEXED WITH CYCLOSPORIN A | | Descriptor: | CYCLOSPORIN A, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE 1 | | Authors: | Ellis, P.J, Carlow, C.K.S, Ma, D, Kuhn, P. | | Deposit date: | 1999-11-22 | | Release date: | 1999-12-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal Structure of the Complex of Brugia Malayi Cyclophilin and Cyclosporin A.
Biochemistry, 39, 2000
|
|
1C5H
 
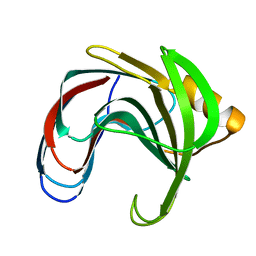 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
1C5I
 
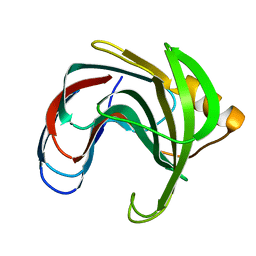 | | HYDROGEN BONDING AND CATALYSIS: AN UNEXPECTED EXPLANATION FOR HOW A SINGLE AMINO ACID SUBSTITUTION CAN CHANGE THE PH OPTIMUM OF A GLYCOSIDASE | | Descriptor: | ENDO-1,4-BETA-XYLANASE, beta-D-xylopyranose-(1-4)-1,5-anhydro-2-deoxy-2-fluoro-D-xylitol | | Authors: | Joshi, M.D, Sidhu, G, Pot, I, Brayer, G.D, Withers, S.G, Mcintosh, L.P. | | Deposit date: | 1999-11-24 | | Release date: | 2000-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase.
J.Mol.Biol., 299, 2000
|
|
1C5L
 
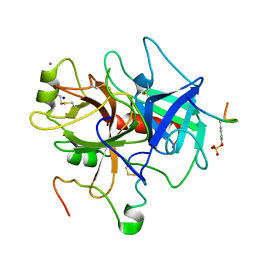 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5M
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | PROTEIN (COAGULATION FACTOR X) | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5N
 
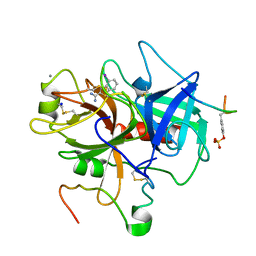 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, Hirudin, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5O
 
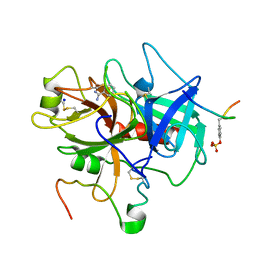 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZAMIDINE, Hirudin, SODIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5P
 
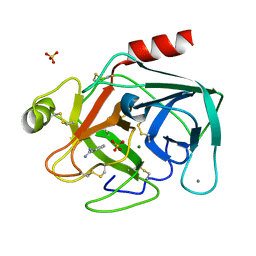 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZAMIDINE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5Q
 
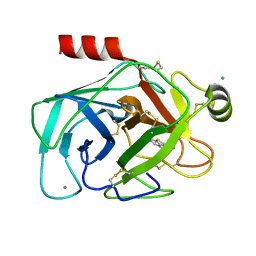 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5R
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | 4-IODOBENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5S
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | BENZO[B]THIOPHENE-2-CARBOXAMIDINE, CALCIUM ION, PROTEIN (TRYPSIN), ... | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
1C5T
 
 | | STRUCTURAL BASIS FOR SELECTIVITY OF A SMALL MOLECULE, S1-BINDING, SUB-MICROMOLAR INHIBITOR OF UROKINASE TYPE PLASMINOGEN ACTIVATOR | | Descriptor: | CALCIUM ION, PROTEIN (TRYPSIN), THIENO[2,3-B]PYRIDINE-2-CARBOXAMIDINE | | Authors: | Katz, B.A, Mackman, R, Luong, C, Radika, K, Martelli, A, Sprengeler, P.A, Wang, J, Chan, H, Wong, L. | | Deposit date: | 1999-12-22 | | Release date: | 2000-12-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural basis for selectivity of a small molecule, S1-binding, submicromolar inhibitor of urokinase-type plasminogen activator.
Chem.Biol., 7, 2000
|
|
