1EZ1
 
 | | STRUCTURE OF ESCHERICHIA COLI PURT-ENCODED GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE COMPLEXED WITH MG, AMPPNP, AND GAR | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ACETATE ION, GLYCINAMIDE RIBONUCLEOTIDE, ... | | Authors: | Thoden, J.B, Firestine, S, Nixon, A, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2000-05-09 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular structure of Escherichia coli PurT-encoded glycinamide ribonucleotide transformylase.
Biochemistry, 39, 2000
|
|
1EZ3
 
 | |
1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
1EZF
 
 | | CRYSTAL STRUCTURE OF HUMAN SQUALENE SYNTHASE | | Descriptor: | FARNESYL-DIPHOSPHATE FARNESYLTRANSFERASE, N-{2-[TRANS-7-CHLORO-1-(2,2-DIMETHYL-PROPYL) -5-NAPHTHALEN-1-YL-2-OXO-1,2,3,5-TETRAHYDRO-BENZO[E] [1,4]OXAZEPIN-3-YL]-ACETYL}-ASPARTIC ACID | | Authors: | Pandit, J, Danley, D.E, Schulte, G.K, Mazzalupo, S.M, Pauly, T.A, Hayward, C.M, Hamanaka, E.S, Thompson, J.F, Harwood, H.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-10-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of human squalene synthase. A key enzyme in cholesterol biosynthesis.
J.Biol.Chem., 275, 2000
|
|
1EZI
 
 | | Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP | | Descriptor: | CMP-N-ACETYLNEURAMINIC ACID SYNTHETASE | | Authors: | Mosimann, S.C, Gilbert, M, Dombrowski, D, Wakarchuk, W, Strynadka, N.C. | | Deposit date: | 2000-05-11 | | Release date: | 2001-02-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a sialic acid-activating synthetase, CMP-acylneuraminate synthetase in the presence and absence of CDP.
J.Biol.Chem., 276, 2001
|
|
1EZJ
 
 | | CRYSTAL STRUCTURE OF THE MULTIMERIZATION DOMAIN OF THE PHOSPHOPROTEIN FROM SENDAI VIRUS | | Descriptor: | CALCIUM ION, ETHYL MERCURY ION, NUCLEOCAPSID PHOSPHOPROTEIN | | Authors: | Tarbouriech, N, Curran, J, Ruigrok, R.W.H, Burmeister, W.P. | | Deposit date: | 2000-05-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tetrameric coiled coil domain of Sendai virus phosphoprotein.
Nat.Struct.Biol., 7, 2000
|
|
1EZK
 
 | | Crystal structure of recombinant tryparedoxin I | | Descriptor: | TRYPAREDOXIN I | | Authors: | Hofmann, B, Guerrero, S.A, Kalisz, H.M, Menge, U, Nogoceke, E, Montemartini, M, Singh, M, Flohe, L, Hecht, H.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of tryparedoxins revealing interaction with trypanothione.
Biol.Chem., 382, 2001
|
|
1EZQ
 
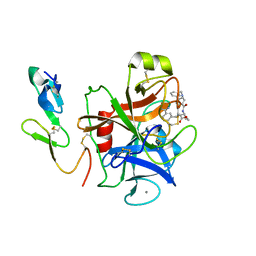 | | CRYSTAL STRUCTURE OF HUMAN COAGULATION FACTOR XA COMPLEXED WITH RPR128515 | | Descriptor: | 3-[(3'-AMINOMETHYL-BIPHENYL-4-CARBONYL)-AMINO]-2-(3-CARBAMIMIDOYL-BENZYL)-BUTYRIC ACID METHYL ESTER, CALCIUM ION, COAGULATION FACTOR XA | | Authors: | Maignan, S, Guilloteau, J.P, Pouzieux, S, Choi-Sledeski, Y.M, Becker, M.R, Klein, S.I, Ewing, W.R, Pauls, H.W, Spada, A.P, Mikol, V. | | Deposit date: | 2000-05-11 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of human factor Xa complexed with potent inhibitors.
J.Med.Chem., 43, 2000
|
|
1EZS
 
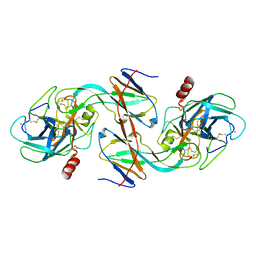 | | CRYSTAL STRUCTURE OF ECOTIN MUTANT M84R, W67A, G68A, Y69A, D70A BOUND TO RAT ANIONIC TRYPSIN II | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZU
 
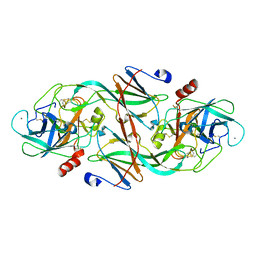 | | ECOTIN Y69F, D70P BOUND TO D102N TRYPSIN | | Descriptor: | CALCIUM ION, ECOTIN, TRYPSIN II, ... | | Authors: | Gillmor, S.A, Takeuchi, T, Yang, S.Q, Craik, C.S, Fletterick, R.J. | | Deposit date: | 2000-05-11 | | Release date: | 2000-06-23 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Compromise and accommodation in ecotin, a dimeric macromolecular inhibitor of serine proteases.
J.Mol.Biol., 299, 2000
|
|
1EZW
 
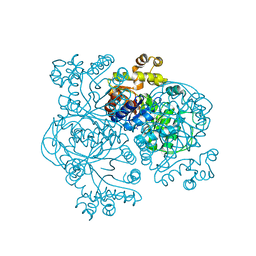 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOPYRUS KANDLERI | | Descriptor: | CHLORIDE ION, COENZYME F420-DEPENDENT N5,N10-METHYLENETETRAHYDROMETHANOPTERIN REDUCTASE, MAGNESIUM ION | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-12 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1EZX
 
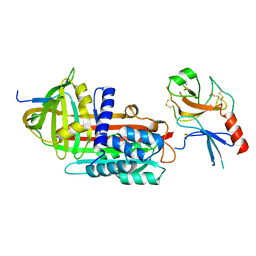 | |
1EZZ
 
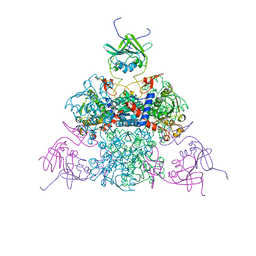 | |
1F00
 
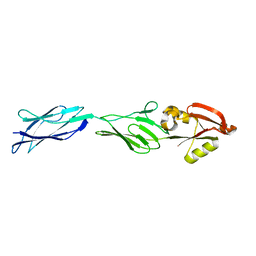 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF ENTEROPATHOGENIC E. COLI INTIMIN | | Descriptor: | INTIMIN | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-12 | | Release date: | 2000-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F02
 
 | | CRYSTAL STRUCTURE OF C-TERMINAL 282-RESIDUE FRAGMENT OF INTIMIN IN COMPLEX WITH TRANSLOCATED INTIMIN RECEPTOR (TIR) INTIMIN-BINDING DOMAIN | | Descriptor: | INTIMIN, TRANSLOCATED INTIMIN RECEPTOR | | Authors: | Luo, Y, Frey, E.A, Pfuetzner, R.A, Creagh, A.L, Knoechel, D.G, Haynes, C.A, Finlay, B.B, Strynadka, N.C.J. | | Deposit date: | 2000-05-14 | | Release date: | 2000-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of enteropathogenic Escherichia coli intimin-receptor complex.
Nature, 405, 2000
|
|
1F05
 
 | |
1F06
 
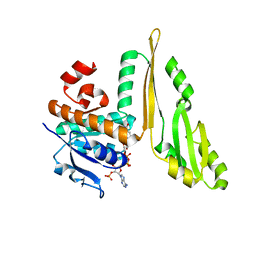 | | THREE DIMENSIONAL STRUCTURE OF THE TERNARY COMPLEX OF CORYNEBACTERIUM GLUTAMICUM DIAMINOPIMELATE DEHYDROGENASE NADPH-L-2-AMINO-6-METHYLENE-PIMELATE | | Descriptor: | L-2-AMINO-6-METHYLENE-PIMELIC ACID, MESO-DIAMINOPIMELATE D-DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cirilli, M, Scapin, G, Sutherland, A, Caplan, J.F, Vederas, J.C, Blanchard, J.S. | | Deposit date: | 2000-05-14 | | Release date: | 2001-05-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of the ternary complex of Corynebacterium glutamicum diaminopimelate dehydrogenase-NADPH-L-2-amino-6-methylene-pimelate.
Protein Sci., 9, 2000
|
|
1F07
 
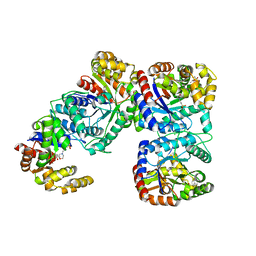 | | STRUCTURE OF COENZYME F420 DEPENDENT TETRAHYDROMETHANOPTERIN REDUCTASE FROM METHANOBACTERIUM THERMOAUTOTROPHICUM | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Shima, S, Warkentin, E, Grabarse, W, Thauer, R.K, Ermler, U. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of coenzyme F(420) dependent methylenetetrahydromethanopterin reductase from two methanogenic archaea.
J.Mol.Biol., 300, 2000
|
|
1F09
 
 | | CRYSTAL STRUCTURE OF THE GREEN FLUORESCENT PROTEIN (GFP) VARIANT YFP-H148Q WITH TWO BOUND IODIDES | | Descriptor: | GREEN FLUORESCENT PROTEIN, IODIDE ION | | Authors: | Wachter, R.M, Yarbrough, D, Kallio, K, Remington, S.J. | | Deposit date: | 2000-05-15 | | Release date: | 2000-11-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystallographic and energetic analysis of binding of selected anions to the yellow variants of green fluorescent protein.
J.Mol.Biol., 301, 2000
|
|
1F0B
 
 | |
1F0C
 
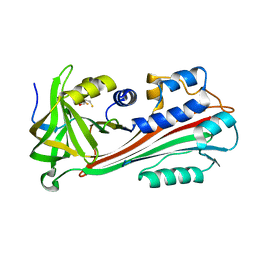 | | STRUCTURE OF THE VIRAL SERPIN CRMA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ICE INHIBITOR | | Authors: | Renatus, M, Zhou, Q, Stennicke, H.R, Snipas, S.J, Turk, D, Bankston, L.A, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2000-05-15 | | Release date: | 2000-09-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal structure of the apoptotic suppressor CrmA in its cleaved form.
Structure Fold.Des., 8, 2000
|
|
1F0K
 
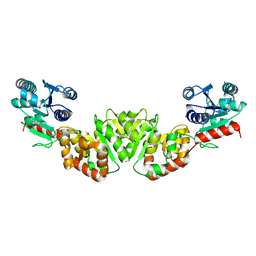 | | THE 1.9 ANGSTROM CRYSTAL STRUCTURE OF E. COLI MURG | | Descriptor: | SULFATE ION, UDP-N-ACETYLGLUCOSAMINE-N-ACETYLMURAMYL-(PENTAPEPTIDE) PYROPHOSPHORYL-UNDECAPRENOL N-ACETYLGLUCOSAMINE TRANSFERASE | | Authors: | Ha, S, Walker, D, Shi, Y, Walker, S. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A crystal structure of Escherichia coli MurG, a membrane-associated glycosyltransferase involved in peptidoglycan biosynthesis.
Protein Sci., 9, 2000
|
|
1F0L
 
 | |
1F0M
 
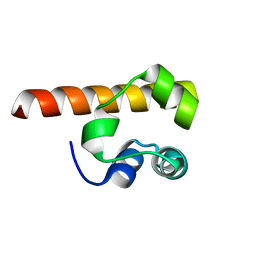 | | MONOMERIC STRUCTURE OF THE HUMAN EPHB2 SAM (STERILE ALPHA MOTIF) DOMAIN | | Descriptor: | EPHRIN TYPE-B RECEPTOR 2 | | Authors: | Thanos, C.D, Faham, S, Goodwill, K.E, Cascio, D, Phillips, M, Bowie, J.U. | | Deposit date: | 2000-05-16 | | Release date: | 2000-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monomeric structure of the human EphB2 sterile alpha motif domain.
J.Biol.Chem., 274, 1999
|
|
1F0N
 
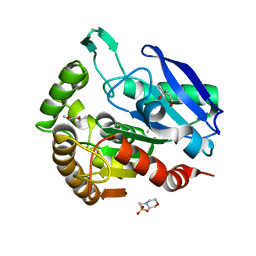 | | MYCOBACTERIUM TUBERCULOSIS ANTIGEN 85B | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ANTIGEN 85B | | Authors: | Anderson, D.H, Harth, G, Horwitz, M.A, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2000-05-16 | | Release date: | 2001-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An interfacial mechanism and a class of inhibitors inferred from two crystal structures of the Mycobacterium tuberculosis 30 kDa major secretory protein (Antigen 85B), a mycolyl transferase.
J.Mol.Biol., 307, 2001
|
|
