1CQD
 
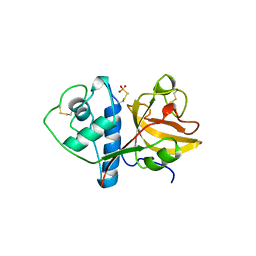 | | THE 2.1 ANGSTROM STRUCTURE OF A CYSTEINE PROTEASE WITH PROLINE SPECIFICITY FROM GINGER RHIZOME, ZINGIBER OFFICINALE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (PROTEASE II), ... | | Authors: | Choi, K.H, Laursen, R.A, Allen, K.N. | | Deposit date: | 1999-06-15 | | Release date: | 1999-09-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 2.1 A structure of a cysteine protease with proline specificity from ginger rhizome, Zingiber officinale.
Biochemistry, 38, 1999
|
|
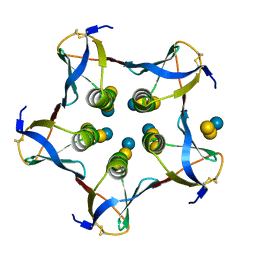
1CQF
 
 | |
1CQI
 
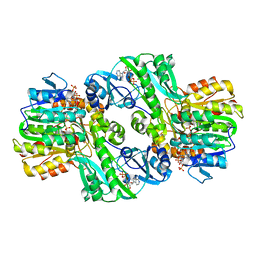 | | Crystal Structure of the Complex of ADP and MG2+ with Dephosphorylated E. Coli Succinyl-CoA Synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, COENZYME A, MAGNESIUM ION, ... | | Authors: | Joyce, M.A, Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 1999-08-06 | | Release date: | 2000-01-07 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography.
Biochemistry, 39, 2000
|
|
1CQJ
 
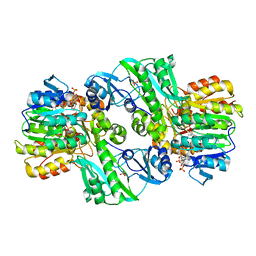 | | CRYSTAL STRUCTURE OF DEPHOSPHORYLATED E. COLI SUCCINYL-COA SYNTHETASE | | Descriptor: | COENZYME A, PHOSPHATE ION, SUCCINYL-COA SYNTHETASE ALPHA CHAIN, ... | | Authors: | Joyce, M.A, Fraser, M.E, James, M.N.G, Bridger, W.A, Wolodko, W.T. | | Deposit date: | 1999-08-06 | | Release date: | 2000-01-10 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | ADP-binding site of Escherichia coli succinyl-CoA synthetase revealed by x-ray crystallography.
Biochemistry, 39, 2000
|
|
1CQK
 
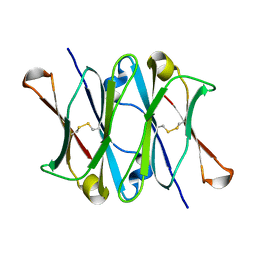 | | CRYSTAL STRUCTURE OF THE CH3 DOMAIN FROM THE MAK33 ANTIBODY | | Descriptor: | CH3 DOMAIN OF MAK33 ANTIBODY | | Authors: | Thies, M.J, Mayer, J, Augustine, J.G, Frederick, C.A, Lilie, H, Buchner, J. | | Deposit date: | 1999-08-06 | | Release date: | 1999-09-11 | | Last modified: | 2018-08-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding and association of the antibody domain CH3: prolyl isomerization preceeds dimerization.
J.Mol.Biol., 293, 1999
|
|
1CQM
 
 | |
1CQN
 
 | |
1CQP
 
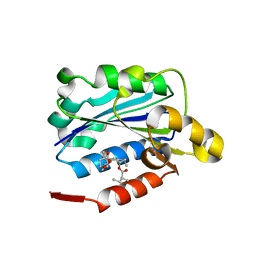 | | CRYSTAL STRUCTURE ANALYSIS OF THE COMPLEX LFA-1 (CD11A) I-DOMAIN / LOVASTATIN AT 2.6 A RESOLUTION | | Descriptor: | ANTIGEN CD11A (P180), LOVASTATIN, MAGNESIUM ION | | Authors: | Kallen, J, Welzenbach, K, Ramage, P, Geyl, D, Kriwacki, R, Legge, G, Cottens, S, Weitz-Schmidt, G, Hommel, U. | | Deposit date: | 1999-08-10 | | Release date: | 2000-08-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for LFA-1 inhibition upon lovastatin binding to the CD11a I-domain.
J.Mol.Biol., 292, 1999
|
|
1CQS
 
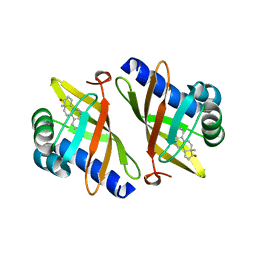 | | CRYSTAL STRUCTURE OF D103E MUTANT WITH EQUILENINEOF KSI IN PSEUDOMONAS PUTIDA | | Descriptor: | EQUILENIN, PROTEIN : KETOSTEROID ISOMERASE | | Authors: | Choi, G, Ha, N.C, Kim, S.W, Kim, D.H, Park, S, Oh, B.H, Choi, K.Y. | | Deposit date: | 1999-08-11 | | Release date: | 2003-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Asp-99 donates a hydrogen bond not to Tyr-14 but to the steroid directly
in the catalytic mechanism of Delta 5-3-ketosteroid isomerase from
Pseudomonas putida biotype B
Biochemistry, 39, 2000
|
|
1CQV
 
 | |
1CQX
 
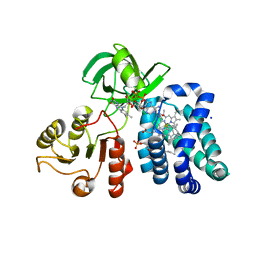 | | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution | | Descriptor: | 1-[GLYCEROLYLPHOSPHONYL]-2-[8-(2-HEXYL-CYCLOPROPYL)-OCTANAL-1-YL]-3-[HEXADECANAL-1-YL]-GLYCEROL, FLAVIN-ADENINE DINUCLEOTIDE, FLAVOHEMOPROTEIN, ... | | Authors: | Ermler, U, Siddiqui, R.A, Cramm, R, Friedrich, B. | | Deposit date: | 1999-08-12 | | Release date: | 1999-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the flavohemoglobin from Alcaligenes eutrophus at 1.75 A resolution.
EMBO J., 14, 1995
|
|
1CR0
 
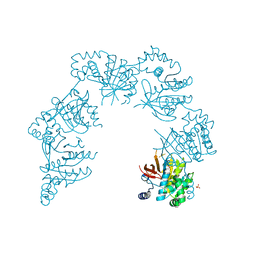 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE4 PROTEIN OF BACTERIOPHAGE T7 | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR1
 
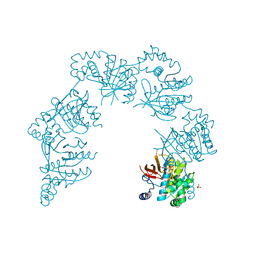 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTTP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR2
 
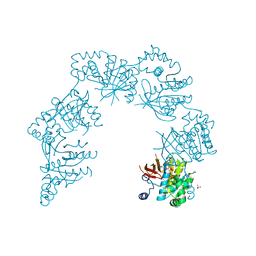 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA PRIMASE/HELICASE, SULFATE ION | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR4
 
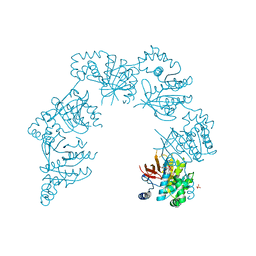 | | CRYSTAL STRUCTURE OF THE HELICASE DOMAIN OF THE GENE 4 PROTEIN OF BACTERIOPHAGE T7: COMPLEX WITH DTDP | | Descriptor: | DNA PRIMASE/HELICASE, SULFATE ION, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Sawaya, M.R, Guo, S, Tabor, S, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1999-08-12 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the helicase domain from the replicative helicase-primase of bacteriophage T7.
Cell(Cambridge,Mass.), 99, 1999
|
|
1CR5
 
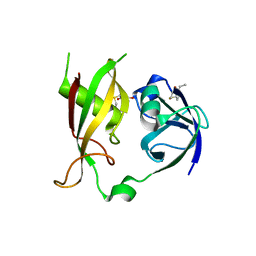 | | N-TERMINAL DOMAIN OF SEC18P | | Descriptor: | 1-ETHYL-PYRROLIDINE-2,5-DIONE, SEC18P (RESIDUES 22 - 210) | | Authors: | Babor, S.M, Fass, D. | | Deposit date: | 1999-08-13 | | Release date: | 1999-12-22 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Sec18p N-terminal domain.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1CR7
 
 | | PEANUT LECTIN-LACTOSE COMPLEX MONOCLINIC FORM | | Descriptor: | CALCIUM ION, LECTIN, MANGANESE (II) ION, ... | | Authors: | Ravishankar, R, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 1999-08-14 | | Release date: | 2001-04-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of the peanut lectin-lactose complex at acidic pH: retention of unusual quaternary structure, empty and carbohydrate bound combining sites, molecular mimicry and crystal packing directed by interactions at the combining site.
Proteins, 43, 2001
|
|
1CRA
 
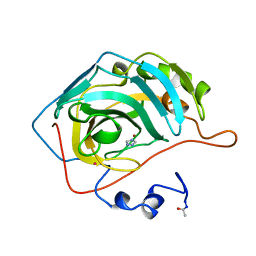 | | THE COMPLEX BETWEEN HUMAN CARBONIC ANHYDRASE II AND THE AROMATIC INHIBITOR 1,2,4-TRIAZOLE | | Descriptor: | 1,2,4-TRIAZOLE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Mangani, S, Liljas, A. | | Deposit date: | 1992-10-21 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the complex between human carbonic anhydrase II and the aromatic inhibitor 1,2,4-triazole.
J.Mol.Biol., 232, 1993
|
|
1CRB
 
 | |
1CRG
 
 | |
1CRH
 
 | |
1CRI
 
 | |
1CRJ
 
 | |
1CRM
 
 | | STRUCTURE AND FUNCTION OF CARBONIC ANHYDRASES | | Descriptor: | CARBONIC ANHYDRASE I, CHLORIDE ION, HYDROSULFURIC ACID, ... | | Authors: | Yadava, V.S, Kannan, K.K. | | Deposit date: | 1994-03-04 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Carbonic Anhydrases
Biomolecular Structure, Conformation, Function and Evolution, 1, 1981
|
|
1CRU
 
 | | SOLUBLE QUINOPROTEIN GLUCOSE DEHYDROGENASE FROM ACINETOBACTER CALCOACETICUS IN COMPLEX WITH PQQ AND METHYLHYDRAZINE | | Descriptor: | CALCIUM ION, GLYCEROL, METHYLHYDRAZINE, ... | | Authors: | Oubrie, A, Rozeboom, H.J, Dijkstra, B.W. | | Deposit date: | 1999-08-16 | | Release date: | 2000-03-01 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Active-site structure of the soluble quinoprotein glucose dehydrogenase complexed with methylhydrazine: a covalent cofactor-inhibitor complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
