1Q79
 
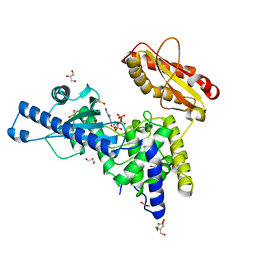 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Martin, G, Moglich, A, Keller, W, Doublie, S. | | Deposit date: | 2003-08-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase.
J.Mol.Biol., 341, 2004
|
|
1Q7B
 
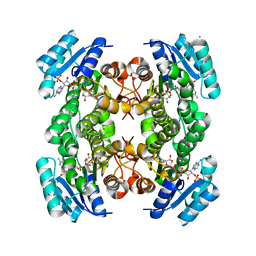 | | The structure of betaketoacyl-[ACP] reductase from E. coli in complex with NADP+ | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, CALCIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Price, A.C, Zhang, Y.-M, Rock, C.O, White, S.M. | | Deposit date: | 2003-08-17 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Cofactor-Induced Conformational Rearrangements Establish a Catalytically Competent Active Site and a Proton Relay Conduit in FabG
Structure, 12, 2004
|
|
1Q7C
 
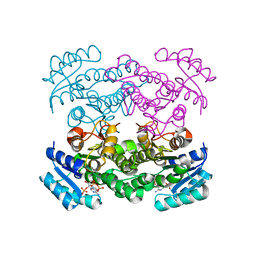 | | The structure of betaketoacyl-[ACP] reductase Y151F mutant in complex with NADPH fragment | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Price, A.C, Zhang, Y.-M, Rock, C.O, White, S.M. | | Deposit date: | 2003-08-17 | | Release date: | 2004-02-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor-Induced Conformational Rearrangements Establish a Catalytically Competent Active Site and a
Proton Relay Conduit in FabG
Structure, 12, 2004
|
|
1Q7D
 
 | |
1Q7E
 
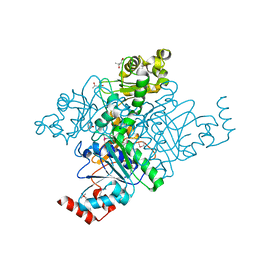 | | Crystal Structure of YfdW protein from E. coli | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Hypothetical protein yfdW, METHIONINE | | Authors: | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-08-18 | | Release date: | 2003-09-30 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1Q7F
 
 | |
1Q7G
 
 | | Homoserine Dehydrogenase in complex with suicide inhibitor complex NAD-5-hydroxy-4-Oxonorvaline | | Descriptor: | Homoserine dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE-5-HYDROXY-4-OXONORVALINE, SODIUM ION | | Authors: | Jacques, S.L, Mirza, I.A, Ejim, L, Koteva, K, Hughes, D.W, Green, K, Kinach, R, Honek, J.F, Lai, H.K, Berghuis, A.M, Wright, G.D. | | Deposit date: | 2003-08-18 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Enzyme assisted suicide: Molecular basis for the antifungal activity of 5-hydroxy-4-oxonorvaline by potent inhibition of homoserine dehydrogenase
Chem.Biol., 10, 2003
|
|
1Q7H
 
 | | Structure of a Conserved PUA Domain Protein from Thermoplasma acidophilum | | Descriptor: | ZINC ION, conserved hypothetical protein | | Authors: | Cuff, M.E, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-08-18 | | Release date: | 2004-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a conserved hypothetical protein from T. acidophilum
TO BE PUBLISHED
|
|
1Q7L
 
 | | Zn-binding domain of the T347G mutant of human aminoacylase-I | | Descriptor: | Aminoacylase-1, GLYCINE, ZINC ION | | Authors: | Lindner, H.A, Lunin, V.V, Alary, A, Hecker, R, Cygler, M, Menard, R. | | Deposit date: | 2003-08-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Essential roles of zinc ligation and enzyme dimerization for catalysis in the aminoacylase-1/M20 family.
J.Biol.Chem., 278, 2003
|
|
1Q7M
 
 | | Cobalamin-dependent methionine synthase (MetH) from Thermotoga maritima (Oxidized, Monoclinic) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q7Q
 
 | | Cobalamin-dependent methionine synthase (1-566) from T. maritima (Oxidized, Orthorhombic) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-19 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q7R
 
 | |
1Q7S
 
 | | Crystal structure of bit1 | | Descriptor: | bit1 | | Authors: | De Pereda, J.M, Waas, W.F, Jan, Y, Ruoslahti, E, Schimmel, P, Pascual, J. | | Deposit date: | 2003-08-19 | | Release date: | 2003-12-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a human peptidyl-tRNA hydrolase reveals a new fold and suggests basis for a bifunctional activity.
J.Biol.Chem., 279, 2004
|
|
1Q7Y
 
 | | Crystal Structure of CCdAP-Puromycin bound at the Peptidyl transferase center of the 50S ribosomal subunit | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q7Z
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q81
 
 | | Crystal Structure of minihelix with 3' puromycin bound to A-site of the 50S ribosomal subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Insights into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q82
 
 | | Crystal Structure of CC-Puromycin bound to the A-site of the 50S ribosomal subunit | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural Insights Into Peptide Bond Formation
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q83
 
 | | Crystal structure of the mouse acetylcholinesterase-TZ2PA6 syn complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Kolb, H.C, Radic, Z, Sharpless, K.B, Taylor, P, Marchot, P. | | Deposit date: | 2003-08-20 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q84
 
 | | Crystal structure of the mouse acetylcholinesterase-TZ2PA6 anti complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-4-YL]HEXYL]-PHENANTHRIDINIUM, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Kolb, H.C, Radic, Z, Sharpless, K.B, Taylor, P, Marchot, P. | | Deposit date: | 2003-08-20 | | Release date: | 2004-02-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Freeze-frame inhibitor captures acetylcholinesterase in a unique conformation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q85
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+ complex, Se-Met) | | Descriptor: | 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q86
 
 | | Crystal structure of CCA-Phe-cap-biotin bound simultaneously at half occupancy to both the A-site and P-site of the the 50S ribosomal Subunit. | | Descriptor: | 23S ribosomal rna, 50S ribosomal protein L13P, 50S ribosomal protein L14P, ... | | Authors: | Hansen, J.L, Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-20 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into peptide bond formation.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1Q87
 
 | |
1Q88
 
 | |
1Q8A
 
 | | Cobalamin-dependent methionine synthase (1-566) from Thermotoga maritima (Cd2+:L-Hcy complex, Se-Met) | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydrofolate S-homocysteine methyltransferase, CADMIUM ION | | Authors: | Evans, J.C, Huddler, D.P, Hilgers, M.T, Romanchuk, G, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 2003-08-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the N-terminal modules imply large domain motions during catalysis by methionine synthase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1Q8B
 
 | |
