1P4B
 
 | | Three-Dimensional Structure Of a Single Chain Fv Fragment Complexed With The peptide GCN4(7P-14P). | | Descriptor: | Antibody Variable heavy chain, Antibody Variable light chain, GCN4(7P-14P) peptide | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-22 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1P4C
 
 | | High Resolution Structure of Oxidized Active Mutant of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-Mandelate Dehydrogenase, ... | | Authors: | Sukumar, N, Mitra, B, Mathews, F.S. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structures of an Oxidized and Reduced Flavoprotein: THE WATER SWITCH IN A SOLUBLE FORM OF (S)-MANDELATE DEHYDROGENASE
J.Biol.Chem., 279, 2004
|
|
1P4D
 
 | | F factor TraI Relaxase Domain | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, TraI protein | | Authors: | Datta, S, Larkin, C, Schildbach, J.F. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into single-stranded DNA binding and cleavage by F factor TraI.
Structure, 11, 2003
|
|
1P4E
 
 | | Flpe W330F mutant-DNA Holliday Junction Complex | | Descriptor: | 33-MER, 5'-D(*TP*AP*AP*GP*TP*TP*CP*CP*TP*AP*TP*TP*C)-3', 5'-D(*TP*TP*TP*AP*AP*AP*AP*GP*AP*AP*TP*AP*GP*GP*AP*AP*CP*TP*TP*C)-3', ... | | Authors: | Rice, P.A, Chen, Y. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The role of the conserved Trp330 in Flp-mediated recombination. Functional and structural analysis
J.Biol.Chem., 278, 2003
|
|
1P4F
 
 | | DEATH ASSOCIATED PROTEIN KINASE CATALYTIC DOMAIN WITH BOUND INHIBITOR FRAGMENT | | Descriptor: | 5,6-Dihydro-benzo[H]cinnolin-3-ylamine, Death-associated protein kinase 1 | | Authors: | Velentza, A.V, Wainwright, M.S, Zasadzki, M, Mirzoeva, S, Haiech, J, Focia, P.J, Egli, M, Watterson, D.M. | | Deposit date: | 2003-04-23 | | Release date: | 2004-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An aminopyridazine-based inhibitor of a pro-apoptotic protein kinase attenuates hypoxia-ischemia induced acute brain injury.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1P4I
 
 | | Crystal Structure of scFv against peptide GCN4 | | Descriptor: | ANTIBODY VARIABLE LIGHT CHAIN, antibody variable heavy chain | | Authors: | Zahnd, C, Spinelli, S, Luginbuhl, B, Jermutus, L, Amstutz, P, Cambillau, C, Pluckthun, A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Directed in Vitro Evolution and Crystallographic Analysis of a Peptide-binding Single Chain Antibody Fragment (scFv) with Low Picomolar Affinity.
J.Biol.Chem., 279, 2004
|
|
1P4K
 
 | |
1P4L
 
 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | Descriptor: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | Authors: | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | Deposit date: | 2003-04-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
1P4M
 
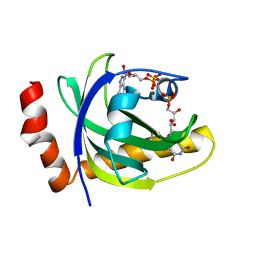 | | CRYSTAL STRUCTURE OF RIBOFLAVIN KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-04-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1P4N
 
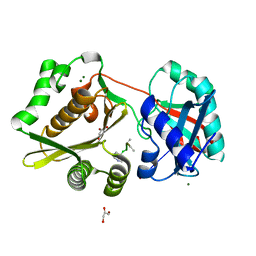 | | Crystal Structure of Weissella viridescens FemX:UDP-MurNAc-pentapeptide complex | | Descriptor: | FemX, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Biarrotte-Sorin, S, Maillard, A, Delettre, J, Sougakoff, W, Arthur, M, Mayer, C. | | Deposit date: | 2003-04-23 | | Release date: | 2004-02-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of Weissella viridescens FemX and its complex with UDP-MurNAc-pentapeptide: insights into FemABX family substrates recognition.
Structure, 12, 2004
|
|
1P4O
 
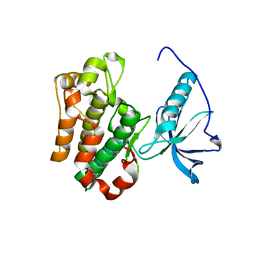 | | Structure of Apo unactivated IGF-1R KInase domain at 1.5A resolution. | | Descriptor: | Insulin-like growth factor I receptor protein | | Authors: | Munshi, S, Kornienko, M, Hall, D.L, Darke, P.L, Waxman, L, Kuo, L.C. | | Deposit date: | 2003-04-23 | | Release date: | 2003-04-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of apo, unactivated insulin-like growth factor-1 receptor kinase at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P4P
 
 | | Outer Surface Protein B of B. burgdorferi: crystal structure of the C-terminal fragment | | Descriptor: | Outer surface protein B | | Authors: | Becker, M, Bunikis, J, Lade, B.D, Dunn, J.J, Barbour, A.G, Lawson, C.L. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-04 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Investigation of Borrelia burgdorferi OspB, a BactericidalFab Target.
J.Biol.Chem., 280, 2005
|
|
1P4R
 
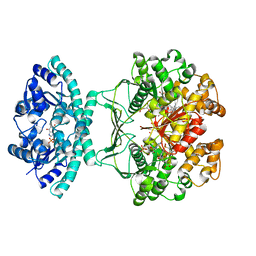 | | Crystal Structure of Human ATIC in complex with folate-based inhibitor BW1540U88UD | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, N-[(S)-(4-{[(2-AMINO-4-HYDROXYQUINAZOLIN-6-YL)(DIHYDROXY)-LAMBDA~4~-SULFANYL]AMINO}PHENYL)(HYDROXY)METHYL]-L-GLUTAMIC ACID, ... | | Authors: | Cheong, C.-G, Greasley, S.E, Horton, P.A, Beardsley, G.P, Wilson, I.A. | | Deposit date: | 2003-04-23 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of human bifunctional enzyme aminoimidazole-4-carboxamide ribonucleotide transformylase/IMP cyclohydrolase in complex with potent sulfonyl-containing antifolates
J.Biol.Chem., 279, 2004
|
|
1P4T
 
 | | Crystal structure of Neisserial surface protein A (NspA) | | Descriptor: | ETHANOLAMINE, PENTAETHYLENE GLYCOL MONODECYL ETHER, SULFATE ION, ... | | Authors: | Vandeputte-Rutten, L, Bos, M.P, Tommassen, J, Gros, P. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Neisserial Surface Protein A (NspA),
a conserved outer membrane protein with vaccine potential
J.Biol.Chem., 278, 2003
|
|
1P4U
 
 | | CRYSTAL STRUCTURE OF GGA3 GAE DOMAIN IN COMPLEX WITH RABAPTIN-5 PEPTIDE | | Descriptor: | ADP-ribosylation factor binding protein GGA3, Rabaptin-5 | | Authors: | Miller, G.J, Mattera, R, Bonifacino, J.S, Hurley, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RECOGNITION OF ACCESSORY PROTEIN MOTIFS BY THE GAMMA-ADAPTIN EAR DOMAIN OF GGA3
Nat.Struct.Biol., 10, 2003
|
|
1P4V
 
 | |
1P4X
 
 | | Crystal structure of SarS protein from Staphylococcus Aureus | | Descriptor: | staphylococcal accessory regulator A homologue | | Authors: | Li, R, Manna, A.C, Dai, S, Cheung, A.L, Zhang, G. | | Deposit date: | 2003-04-24 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the SarS protein from Staphylococcus aureus
J.BACTERIOL., 185, 2003
|
|
1P4Y
 
 | |
1P4Z
 
 | |
1P50
 
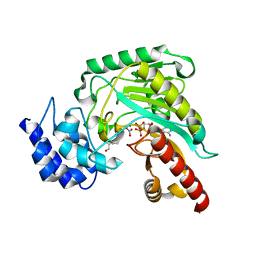 | | Transition state structure of an Arginine Kinase mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P51
 
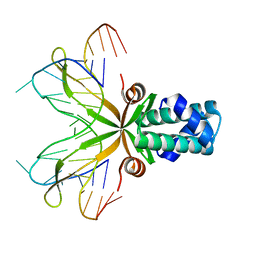 | | Anabaena HU-DNA cocrystal structure (AHU6) | | Descriptor: | 5'-D(*GP*CP*AP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*AP*T)-3', DNA-binding protein HU | | Authors: | Swinger, K.K, Lemberg, K.M, Zhang, Y, Rice, P.A. | | Deposit date: | 2003-04-24 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Flexible DNA bending in HU-DNA cocrystal structures
Embo J., 22, 2003
|
|
1P52
 
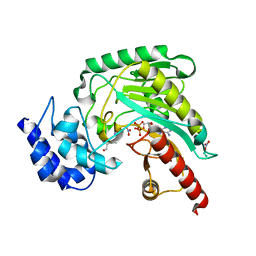 | | Structure of Arginine kinase E314D mutant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, D-ARGININE, ... | | Authors: | Pruett, P.S, Azzi, A, Clark, S.A, Yousef, M.S, Gattis, J.L, Somasundarum, T, Ellington, W.R, Chapman, M.S. | | Deposit date: | 2003-04-24 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase.
J.Biol.Chem., 278, 2003
|
|
1P53
 
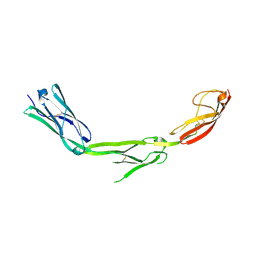 | | The Crystal Structure of ICAM-1 D3-D5 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1 | | Authors: | Yang, Y, Jun, C.D, Liu, J.H, Zhang, R, Jochimiak, A, Springer, T.A, Wang, J.H. | | Deposit date: | 2003-04-24 | | Release date: | 2004-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural basis for dimerization of ICAM-1 on the cell surface.
Mol.Cell, 14, 2004
|
|
1P54
 
 | |
1P56
 
 | |
