1NPF
 
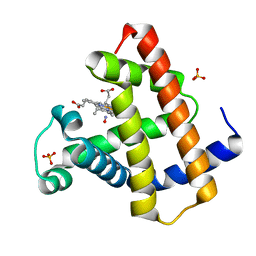 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | Myoglobin, NITRIC OXIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of ferrous horse heart myoglobin complexed with nitric oxide and nitrosoethane.
Proteins, 53, 2003
|
|
1NPG
 
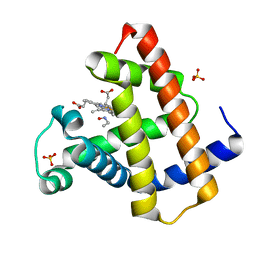 | | MYOGLOBIN (HORSE HEART) WILD-TYPE COMPLEXED WITH NITROSOETHANE | | Descriptor: | Myoglobin, NITROSOETHANE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Copeland, D.M, West, A.H, Richter-Addo, G.B. | | Deposit date: | 2003-01-17 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of ferrous horse heart myoglobin complexed with nitric oxide and nitrosoethane.
Proteins, 53, 2003
|
|
1NPH
 
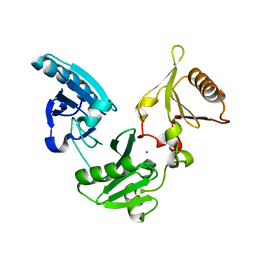 | | Gelsolin Domains 4-6 in Active, Actin Free Conformation Identifies Sites of Regulatory Calcium Ions | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Kolappan, S, Gooch, J.T, Weeds, A.G, McLaughlin, P.J. | | Deposit date: | 2003-01-17 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Gelsolin Domains 4-6 in Active, Actin-Free Conformation Identifies Sites of Regulatory Calcium Ions
J.Mol.Biol., 329, 2003
|
|
1NPI
 
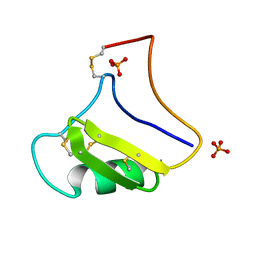 | | Tityus Serrulatus Neurotoxin (Ts1) at atomic resolution | | Descriptor: | PHOSPHATE ION, Toxin VII | | Authors: | Pinheiro, C.B, Marangoni, S, Toyama, M.H, Polikarpov, I. | | Deposit date: | 2003-01-17 | | Release date: | 2003-02-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Structural analysis of Tityus serrulatus Ts1 neurotoxin at atomic resolution: insights into interactions with Na+ channels.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NPJ
 
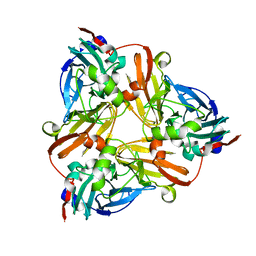 | | Crystal structure of H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reconstitution of the type-1 active site of the H145G/A variants of nitrite reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1NPL
 
 | | MANNOSE-SPECIFIC AGGLUTININ (LECTIN) FROM DAFFODIL (NARCISSUS PSEUDONARCISSUS) BULBS IN COMPLEX WITH MANNOSE-ALPHA1,3-MANNOSE | | Descriptor: | PHOSPHATE ION, PROTEIN (AGGLUTININ), alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose | | Authors: | Sauerborn, M.K, Wright, L.M, Reynolds, C.D, Grossmann, J.G, Rizkallah, P.J. | | Deposit date: | 1998-12-17 | | Release date: | 1998-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into carbohydrate recognition by Narcissus pseudonarcissus lectin: the crystal structure at 2 A resolution in complex with alpha1-3 mannobiose.
J.Mol.Biol., 290, 1999
|
|
1NPN
 
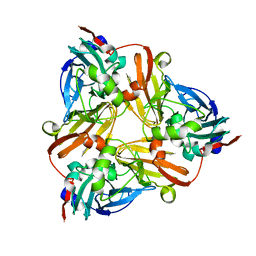 | | Crystal structure of a copper reconstituted H145A mutant of nitrite reductase from Alcaligenes faecalis | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Wijma, H.J, Boulanger, M.J, Molon, A, Fittipaldi, M, Huber, M, Murphy, M.E, Verbeet, M.P, Canters, G.W. | | Deposit date: | 2003-01-18 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reconstitution of the type-1 active site of the 145G/A variants of Nitrite Reductase by ligand insertion
Biochemistry, 42, 2003
|
|
1NPP
 
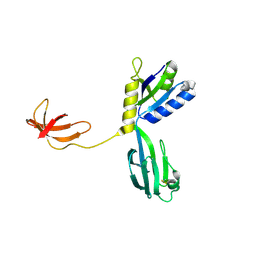 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN P2(1) | | Descriptor: | ISOPROPYL ALCOHOL, Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzahn, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1NPR
 
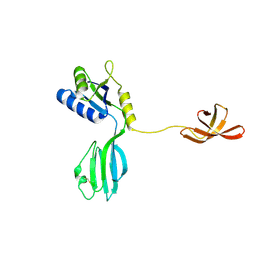 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1NPT
 
 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 replaced by Ala complexed with NAD+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NPU
 
 | | CRYSTAL STRUCTURE OF THE EXTRACELLULAR DOMAIN OF MURINE PD-1 | | Descriptor: | Programmed cell death protein 1 | | Authors: | Zhang, X, Schwartz, J.-C.D, Guo, X, Cao, E, Chen, L, Zhang, Z.-Y, Nathenson, S.G, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-20 | | Release date: | 2004-03-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional analysis of the costimulatory receptor programmed death-1.
Immunity, 20, 2004
|
|
1NPX
 
 | | STRUCTURE OF NADH PEROXIDASE FROM STREPTOCOCCUS FAECALIS 10C1 REFINED AT 2.16 ANGSTROMS RESOLUTION | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH PEROXIDASE | | Authors: | Stehle, T, Ahmed, S.A, Claiborne, A, Schulz, G.E. | | Deposit date: | 1991-08-02 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure of NADH peroxidase from Streptococcus faecalis 10C1 refined at 2.16 A resolution.
J.Mol.Biol., 221, 1991
|
|
1NPY
 
 | | Structure of shikimate 5-dehydrogenase-like protein HI0607 | | Descriptor: | ACETYL GROUP, Hypothetical shikimate 5-dehydrogenase-like protein HI0607 | | Authors: | Korolev, S, Koroleva, O, Zarembinski, T, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-20 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of a Novel Shikimate Dehydrogenase from Haemophilus influenzae.
J.Biol.Chem., 280, 2005
|
|
1NPZ
 
 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N~2~-(morpholin-4-ylcarbonyl)-N-[(3S)-1-phenyl-5-(phenylsulfonyl)pentan-3-yl]-L-leucinamide | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Seddon, A.P, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
1NQ3
 
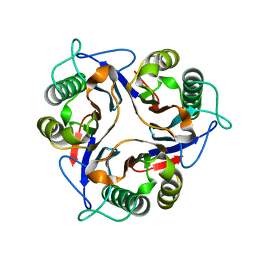 | | Crystal structure of the mammalian tumor associated antigen UK114 | | Descriptor: | 14.3 kDa perchloric acid soluble protein | | Authors: | Deriu, D, Briand, C, Mistiniene, E, Naktinis, V, Grutter, M.G. | | Deposit date: | 2003-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and oligomeric state of the mammalian tumour-associated antigen UK114.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NQ5
 
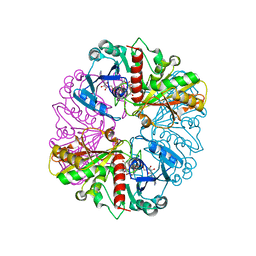 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ser Complexed With Nad+ | | Descriptor: | Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQ6
 
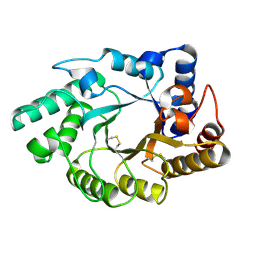 | | Crystal Structure of the catalytic domain of xylanase A from Streptomyces halstedii JM8 | | Descriptor: | MAGNESIUM ION, Xys1 | | Authors: | Canals, A, Vega, M.C, Gomis-Ruth, F.X, Santamaria, R.I, Coll, M. | | Deposit date: | 2003-01-21 | | Release date: | 2004-01-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of xylanase Xys1delta from Streptomyces halstedii.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NQ7
 
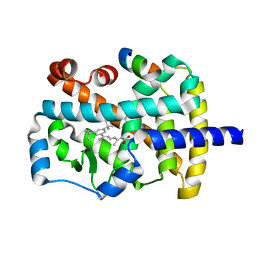 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | 7-(3,5-DITERT-BUTYLPHENYL)-3-METHYLOCTA-2,4,6-TRIENOIC ACID, NUCLEAR RECEPTOR ROR-BETA, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
1NQ9
 
 | | Crystal Structure of Antithrombin in the Pentasaccharide-Bound Intermediate State | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,4-di-O-methyl-2,6-di-O-sulfo-alpha-D-glucopyranose-(1-4)-2,3-di-O-methyl-beta-D-glucopyranuronic acid-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-3-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2,3,6-tri-O-sulfo-alpha-D-glucopyranoside, ... | | Authors: | Huntington, J.A, Johnson, D.J.D. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Antithrombin in a Heparin-Bound Intermediate State
Biochemistry, 42, 2003
|
|
1NQA
 
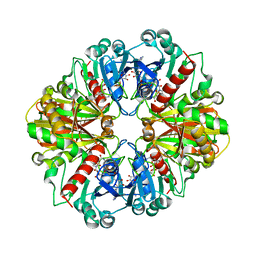 | | Glyceraldehyde-3-Phosphate Dehydrogenase Mutant With Cys 149 Replaced By Ala Complexed With Nad+ and D-Glyceraldehyde-3-Phosphate | | Descriptor: | GLYCERALDEHYDE-3-PHOSPHATE, Glyceraldehyde 3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Didierjean, C, Corbier, C, Fatih, M, Favier, F, Boschi-Muller, S, Branlant, G, Aubry, A. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of two ternary complexes of phosphorylating
Glyceraldehyde-3-Phosphate Dehydrogenase from Bacillus stearothermophilus
with NAD and D-Glyceraldehyde-3-Phosphate
J.Biol.Chem., 278, 2003
|
|
1NQB
 
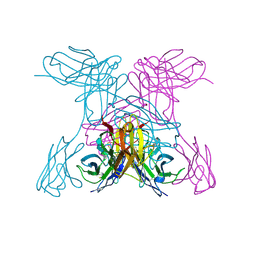 | | TRIVALENT ANTIBODY FRAGMENT | | Descriptor: | SINGLE-CHAIN ANTIBODY FRAGMENT | | Authors: | Williams, R.L, Pei, X.Y. | | Deposit date: | 1997-04-04 | | Release date: | 1997-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0-A resolution crystal structure of a trimeric antibody fragment with noncognate VH-VL domain pairs shows a rearrangement of VH CDR3.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1NQC
 
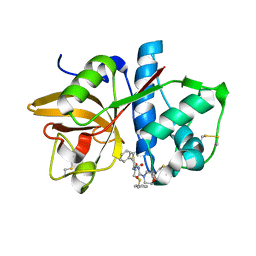 | | Crystal structures of Cathepsin S inhibitor complexes | | Descriptor: | Cathepsin S, N-[(1R)-2-(BENZYLSULFANYL)-1-FORMYLETHYL]-N-(MORPHOLIN-4-YLCARBONYL)-L-PHENYLALANINAMIDE | | Authors: | Pauly, T.A, Sulea, T, Ammirati, M, Sivaraman, J, Danley, D.E, Griffor, M.C, Kamath, A.V, Wang, I.K, Laird, E.R, Menard, R, Cygler, M, Rath, V.L. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity determinants of human cathepsin s revealed
by crystal structures of complexes.
Biochemistry, 42, 2003
|
|
1NQD
 
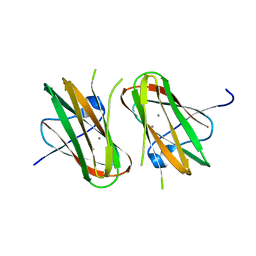 | | CRYSTAL STRUCTURE OF CLOSTRIDIUM HISTOLYTICUM COLG COLLAGENASE COLLAGEN-BINDING DOMAIN 3B AT 1.65 ANGSTROM RESOLUTION IN PRESENCE OF CALCIUM | | Descriptor: | CALCIUM ION, class 1 collagenase | | Authors: | Wilson, J.J, Matsushita, O, Okabe, A, Sakon, J. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Bacterial Collagen-Binding Domain with Novel Calcium-Binding Motif Controls Domain Orientation
Embo J., 22, 2003
|
|
1NQE
 
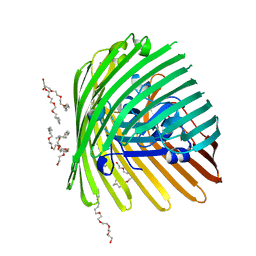 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, Vitamin B12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
1NQF
 
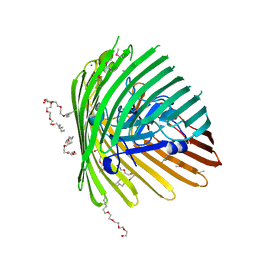 | | OUTER MEMBRANE COBALAMIN TRANSPORTER (BTUB) FROM E. COLI, METHIONINE SUBSTIUTION CONSTRUCT FOR SE-MET SAD PHASING | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, Vitamin B12 receptor | | Authors: | Chimento, D.P, Mohanty, A.K, Kadner, R.J, Wiener, M.C. | | Deposit date: | 2003-01-21 | | Release date: | 2003-04-29 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate-induced transmembrane signaling in the cobalamin transporter BtuB
Nat.Struct.Biol., 10, 2003
|
|
