1MHH
 
 | |
1MHN
 
 | |
1MHO
 
 | | THE 2.0 A STRUCTURE OF HOLO S100B FROM BOVINE BRAIN | | Descriptor: | CALCIUM ION, S-100 PROTEIN | | Authors: | Matsumura, H, Shiba, T, Inoue, T, Harada, S, Yasushi, K.A.I. | | Deposit date: | 1997-09-11 | | Release date: | 1998-11-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mode of target recognition suggested by the 2.0 A structure of holo S100B from bovine brain.
Structure, 6, 1998
|
|
1MHP
 
 | | Crystal structure of a chimeric alpha1 integrin I-domain in complex with the Fab fragment of a humanized neutralizing antibody | | Descriptor: | FAB FRAGMENT, light chain, Fab fragment, ... | | Authors: | Karpusas, M, Taylor, F, Ferrant, J, Weinreb, P, Garber, E. | | Deposit date: | 2002-08-20 | | Release date: | 2003-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the alpha 1 beta 1 Integrin I Domain in Complex with
an Antibody Fab Fragment
J.Mol.Biol., 327, 2003
|
|
1MHQ
 
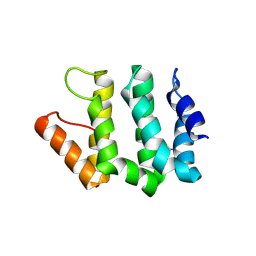 | | Crystal Structure Of Human GGA2 VHS Domain | | Descriptor: | ADP-ribosylation factor binding protein GGA2 | | Authors: | Zhu, G, Zhang, X.C. | | Deposit date: | 2002-08-20 | | Release date: | 2003-03-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of GGA2 VHS domain and its implication in plasticity in the ligand binding pocket
FEBS LETT., 537, 2003
|
|
1MHW
 
 | | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides | | Descriptor: | 4-biphenylacetyl-Cys-(D)Arg-Tyr-N-(2-phenylethyl) amide, Cathepsin L | | Authors: | Chowdhury, S, Sivaraman, J, Wang, J, Devanathan, G, Lachance, P, Qi, H, Menard, R, Lefebvre, J, Konishi, Y, Cygler, M, Sulea, T, Purisima, E.O. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design of non-covalent inhibitors of human cathepsin L. From the 96-residue proregion to optimized tripeptides
J.Med.Chem., 45, 2002
|
|
1MHX
 
 | | Crystal Structures of the redesigned protein G variant NuG1 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Protein Sci., 11, 2002
|
|
1MI0
 
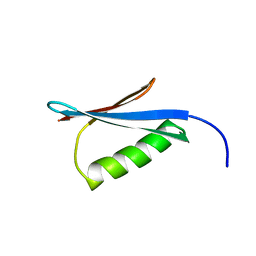 | | Crystal Structure of the redesigned protein G variant NuG2 | | Descriptor: | immunoglobulin-binding protein G | | Authors: | Nauli, S, Kuhlman, B, Le Trong, I, Stenkamp, R.E, Teller, D.C, Baker, D. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures and increased stabilization of the protein G variants with switched folding pathways NuG1 and NuG2
Biochemistry, 11, 2002
|
|
1MI1
 
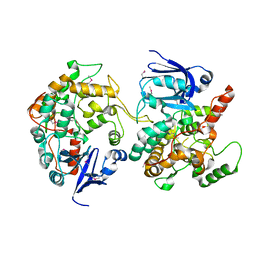 | | Crystal Structure of the PH-BEACH Domain of Human Neurobeachin | | Descriptor: | Neurobeachin | | Authors: | Jogl, G, Shen, Y, Gebauer, D, Li, J, Wiegmann, K, Kashkar, H, Kroenke, M, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the BEACH domain reveals an unusual fold and extensive association with a novel PH domain.
EMBO J., 21, 2002
|
|
1MI3
 
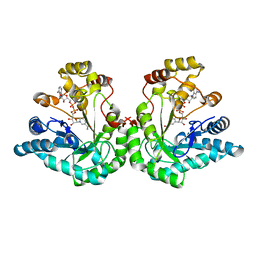 | | 1.8 Angstrom structure of xylose reductase from Candida tenuis in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-08-21 | | Release date: | 2003-08-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of xylose reductase bound to NAD+ and the basis for single and dual co-substrate specificity in family 2 aldo-keto reductases
Biochem.J., 373, 2003
|
|
1MI4
 
 | | Glyphosate insensitive G96A mutant EPSP synthase liganded with shikimate-3-phosphate | | Descriptor: | 5-enolpyruvylshikimate-3-phosphate synthase, FORMIC ACID, SHIKIMATE-3-PHOSPHATE | | Authors: | Eschenburg, S, Healy, M.L, Priestman, M.A, Lushington, G.H, Schonbrunn, E. | | Deposit date: | 2002-08-21 | | Release date: | 2002-12-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | How the mutation glycine96 to alanine confers glyphosate insensitivity to 5-enolpyruvyl
shikimate-3-phosphate synthase from Escherichia coli.
PLANTA, 216, 2002
|
|
1MI7
 
 | | Crystal Structure of Domain Swapped trp Aporepressor in 30%(v/v) Isopropanol | | Descriptor: | ISOPROPYL ALCOHOL, Trp operon repressor | | Authors: | Lawson, C.L, Benoff, B, Berger, T, Berman, H.M, Carey, J. | | Deposit date: | 2002-08-22 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli trp repressor forms a domain-swapped array in aqueous alcohol.
Structure, 12, 2004
|
|
1MI8
 
 | | 2.0 Angstrom crystal structure of a DnaB intein from Synechocystis sp. PCC 6803 | | Descriptor: | DnaB intein | | Authors: | Ding, Y, Chen, X, Ferrandon, S, Xu, M, Rao, Z. | | Deposit date: | 2002-08-22 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of mini-intein reveals a conserved catalytic module involved in side chain cyclization of asparagine during protein splicing
J.Biol.Chem., 278, 2003
|
|
1MID
 
 | |
1MIE
 
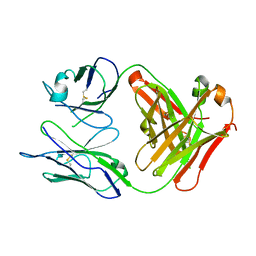 | | Crystal Structure Of The Fab Fragment of Esterolytic Antibody MS5-393 | | Descriptor: | IMMUNOGLOBULIN MS5-393 | | Authors: | Ruzheinikov, S.N, Muranova, T.A, Sedelnikova, S.E, Partridge, L.J, Blackburn, G.M, Murray, I.A, Kakinuma, H, Takashi, N, Shimazaki, K, Sun, J, Nishi, Y, Rice, D.W. | | Deposit date: | 2002-08-23 | | Release date: | 2003-09-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution crystal structure of the Fab-fragments of a family of mouse catalytic antibodies with esterase activity
J.Mol.Biol., 332, 2003
|
|
1MIH
 
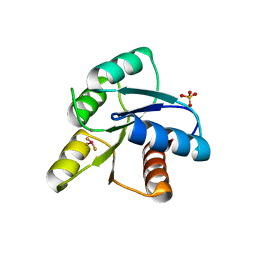 | | A ROLE FOR CHEY GLU 89 IN CHEZ-MEDIATED DEPHOSPHORYLATION OF THE E. COLI CHEMOTAXIS RESPONSE REGULATOR CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, MANGANESE (II) ION, ... | | Authors: | Silversmith, R.E, Guanga, G.P, Betts, L, Chu, C, Zhao, R, Bourret, R.B. | | Deposit date: | 2002-08-23 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | CheZ-mediated dephosphorylation of the Escherichia coli chemotaxis response regulator CheY: role for CheY glutamate 89.
J.Bacteriol., 185, 2003
|
|
1MIJ
 
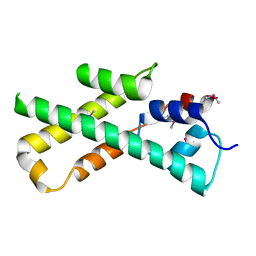 | |
1MIM
 
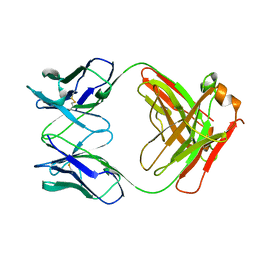 | | IGG FAB FRAGMENT (CD25-BINDING) | | Descriptor: | CHIMERIC SDZ CHI621 | | Authors: | Mikol, V. | | Deposit date: | 1995-12-04 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the fab fragment of SDZ CHI621: a chimeric antibody against CD25.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1MIQ
 
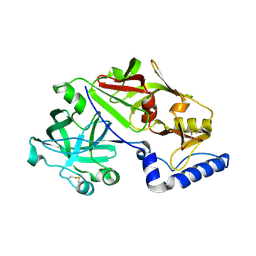 | | Crystal structure of proplasmepsin from the human malarial pathogen Plasmodium vivax | | Descriptor: | plasmepsin | | Authors: | Bernstein, N.K, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N. | | Deposit date: | 2002-08-23 | | Release date: | 2002-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
1MIR
 
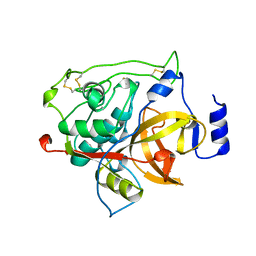 | | RAT PROCATHEPSIN B | | Descriptor: | PROCATHEPSIN B | | Authors: | Cygler, M, Sivaraman, J, Grochulski, P, Coulombe, R, Storer, A.C, Mort, J.S. | | Deposit date: | 1996-01-12 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion.
Structure, 4, 1996
|
|
1MIV
 
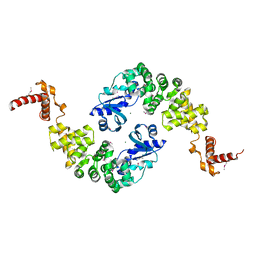 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme | | Descriptor: | MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIX
 
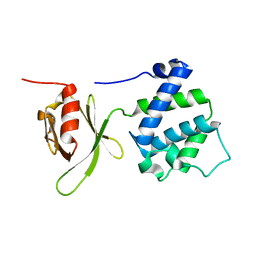 | | Crystal structure of a FERM domain of Talin | | Descriptor: | Talin | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIZ
 
 | | Crystal structure of an integrin beta3-talin chimera | | Descriptor: | TALIN, integrin beta3 | | Authors: | Garcia-Alvarez, B, de Pereda, J.M, Calderwood, D.A, Ulmer, T.S, Critchley, D, Campbell, I.D, Ginsberg, M.H, Liddington, R.C. | | Deposit date: | 2002-08-23 | | Release date: | 2003-01-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural determinants of integrin recognition by talin
Mol.Cell, 11, 2003
|
|
