1K6R
 
 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH MOXALACTAM | | Descriptor: | (2R)-2-((R)-CARBOXY{[CARBOXY(4-HYDROXYPHENYL)ACETYL]AMINO}METHOXYMETHYL)-5-METHYLENE-5,6-DIHYDRO-2H-1,3-OXAZINE-4-CARBO XYLIC ACID, Beta-lactamase PSE-2 | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1K6S
 
 | | STRUCTURE OF THE CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH A PHENYLBORONIC ACID | | Descriptor: | 4-IODO-ACETAMIDO PHENYLBORONIC ACID, Beta-lactamase PSE-2, CALCIUM ION, ... | | Authors: | Kerff, F, Fonze, E, Sauvage, E, Frere, J.M, Charlier, P. | | Deposit date: | 2001-10-17 | | Release date: | 2003-06-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | CRYSTAL STRUCTURE OF CLASS D BETA-LACTAMASE OXA-10 IN COMPLEX WITH DIFFERENT SUBSTRATES AND ONE INHIBITOR.
To be Published
|
|
1K6U
 
 | | Crystal Structure of Cyclic Bovine Pancreatic Trypsin Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Botos, I, Wu, Z, Lu, W, Wlodawer, A. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of a cyclic form of bovine pancreatic trypsin inhibitor.
FEBS Lett., 509, 2001
|
|
1K6W
 
 | | The Structure of Escherichia coli Cytosine Deaminase | | Descriptor: | Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K6X
 
 | | Crystal structure of Nmra, a negative transcriptional regulator in complex with NAD at 1.5 A resolution (Trigonal form) | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NmrA | | Authors: | Stammers, D.K, Ren, J, Leslie, K, Nichols, C.E, Lamb, H.K, Cocklin, S, Dodds, A, Hawkins, A.R. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases.
EMBO J., 20, 2002
|
|
1K6Y
 
 | | Crystal Structure of a Two-Domain Fragment of HIV-1 Integrase | | Descriptor: | Integrase, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, J, Ling, H, Yang, W, Craigie, R. | | Deposit date: | 2001-10-17 | | Release date: | 2001-12-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein.
EMBO J., 20, 2001
|
|
1K6Z
 
 | | Crystal Structure of the Yersinia Secretion Chaperone SycE | | Descriptor: | IMIDAZOLE, Type III secretion chaperone SycE | | Authors: | Evdokimov, A.G, Tropea, J.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2001-10-17 | | Release date: | 2001-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of the type III secretion chaperone SycE from Yersinia pestis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1K70
 
 | | The Structure of Escherichia coli Cytosine Deaminase bound to 4-Hydroxy-3,4-Dihydro-1H-Pyrimidin-2-one | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE, Cytosine Deaminase, FE (III) ION | | Authors: | Ireton, G.C, McDermott, G, Black, M.E, Stoddard, B.L. | | Deposit date: | 2001-10-17 | | Release date: | 2002-02-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of Escherichia coli cytosine deaminase.
J.Mol.Biol., 315, 2002
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | Authors: | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1K75
 
 | | The L-histidinol dehydrogenase (hisD) structure implicates domain swapping and gene duplication. | | Descriptor: | GLYCEROL, L-histidinol dehydrogenase, SULFATE ION | | Authors: | Barbosa, J.A.R.G, Sivaraman, J, Li, Y, Larocque, R, Matte, A, Schrag, J, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2001-10-18 | | Release date: | 2002-02-27 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mechanism of action and NAD+-binding mode revealed by the crystal structure of L-histidinol dehydrogenase.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K77
 
 | | Crystal Structure of EC1530, a Putative Oxygenase from Escherichia coli | | Descriptor: | FORMIC ACID, GLYCEROL, Hypothetical protein ygbM, ... | | Authors: | Kim, Y, Skarina, T, Beasley, S, Laskowski, R, Arrowsmith, C.H, Joachimiak, A, Edwards, A.M, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-10-18 | | Release date: | 2002-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of Escherichia coli EC1530, a glyoxylate induced protein YgbM.
Proteins, 48, 2002
|
|
1K78
 
 | | Pax5(1-149)+Ets-1(331-440)+DNA | | Descriptor: | C-ets-1 Protein, Paired Box Protein Pax5, Pax5/Ets Binding Site on the mb-1 promoter | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K79
 
 | | Ets-1(331-440)+GGAA duplex | | Descriptor: | C-ets-1 protein, DNA (5'-D(*CP*AP*CP*AP*TP*TP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*AP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K7A
 
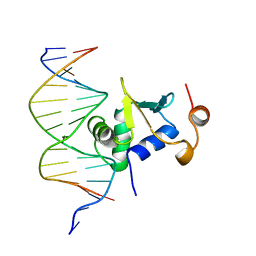 | | Ets-1(331-440)+GGAG duplex | | Descriptor: | C-ets-1 Protein, DNA (5'-D(*CP*AP*CP*AP*TP*CP*TP*CP*CP*GP*GP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*CP*CP*GP*GP*AP*GP*AP*TP*GP*T)-3') | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1K7C
 
 | |
1K7E
 
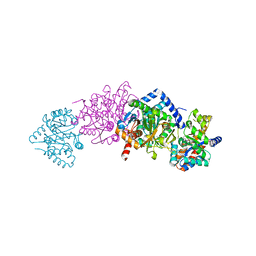 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K7F
 
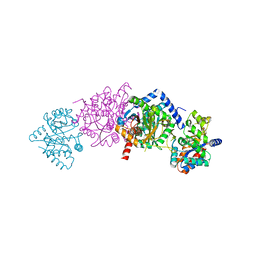 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]VALINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]VALINE ACID, PYRIDOXAL-5'-PHOSPHATE, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
1K7H
 
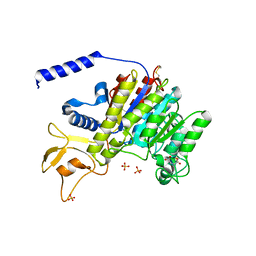 | | CRYSTAL STRUCTURE OF SHRIMP ALKALINE PHOSPHATASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALKALINE PHOSPHATASE, MALEIC ACID, ... | | Authors: | De Backer, M.E, Mc Sweeney, S, Rasmussen, H.B, Riise, B.W, Lindley, P, Hough, E. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The 1.9 A Crystal Structure of Heat-Labile Shrimp Alkaline Phosphatase
J.Mol.Biol., 318, 2002
|
|
1K7T
 
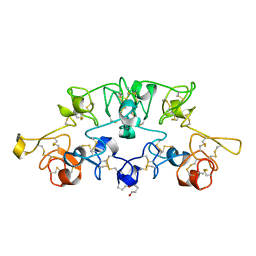 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Gal complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7U
 
 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,4GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7V
 
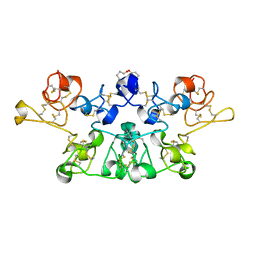 | | Crystal Structure Analysis of crosslinked-WGA3/GlcNAcbeta1,6Galbeta1,4Glc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, agglutinin isolectin 3 | | Authors: | Muraki, M, Ishimura, M, Harata, K. | | Deposit date: | 2001-10-22 | | Release date: | 2002-04-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interactions of wheat-germ agglutinin with GlcNAc beta 1,6Gal sequence
Biochim.Biophys.Acta, 1569, 2002
|
|
1K7W
 
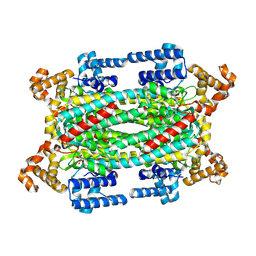 | | Crystal Structure of S283A Duck Delta 2 Crystallin Mutant | | Descriptor: | ARGININOSUCCINATE, delta 2 crystallin | | Authors: | Sampaleanu, L.M, Yu, B, Howell, P.L. | | Deposit date: | 2001-10-22 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Mutational analysis of duck delta 2 crystallin and the structure of an inactive mutant with bound substrate provide insight into the enzymatic mechanism of argininosuccinate lyase.
J.Biol.Chem., 277, 2002
|
|
1K7X
 
 | | CRYSTAL STRUCTURE OF THE BETA-SER178PRO MUTANT OF TRYPTOPHAN SYNTHASE | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, TRYPTOPHAN SYNTHASE ALPHA CHAIN, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the beta Ser178--> Pro mutant of tryptophan synthase. A "knock-out" allosteric enzyme.
J.Biol.Chem., 277, 2002
|
|
1K83
 
 | | Crystal Structure of Yeast RNA Polymerase II Complexed with the Inhibitor Alpha Amanitin | | Descriptor: | ALPHA AMANITIN, DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, ... | | Authors: | Bushnell, D.A, Cramer, P, Kornberg, R.D. | | Deposit date: | 2001-10-22 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Transcription: Alpha-Amanitin-RNA Polymerase II Cocrystal at 2.8 A Resolution.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1K8A
 
 | | Co-crystal structure of Carbomycin A bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
