1IKP
 
 | | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKQ
 
 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
1IKT
 
 | |
1IKV
 
 | | K103N Mutant HIV-1 Reverse Transcriptase in Complex with Efivarenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKW
 
 | | Wild Type HIV-1 Reverse Transcriptase in Complex with Efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IKX
 
 | |
1IKY
 
 | | HIV-1 Reverse Transcriptase in Complex with the Inhibitor MSC194 | | Descriptor: | 1-[2-(3-ACETYL-2-HYDROXY-6-METHOXY-PHENYL)-CYCLOPROPYL]-3-(5-CYANO-PYRIDIN-2-YL)-THIOUREA, POL POLYPROTEIN | | Authors: | Lindberg, J, Unge, T. | | Deposit date: | 2001-05-07 | | Release date: | 2001-06-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the inhibitory efficacy of efavirenz (DMP-266), MSC194 and PNU142721 towards the HIV-1 RT K103N mutant.
Eur.J.Biochem., 269, 2002
|
|
1IL0
 
 | | X-RAY CRYSTAL STRUCTURE OF THE E170Q MUTANT OF HUMAN L-3-HYDROXYACYL-COA DEHYDROGENASE | | Descriptor: | 3-hydroxyacyl-CoA dehydrogenase, ACETOACETYL-COENZYME A, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Barycki, J.J, O'Brien, L.K, Strauss, A.W, Banaszak, L.J. | | Deposit date: | 2001-05-07 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glutamate 170 of human l-3-hydroxyacyl-CoA dehydrogenase is required for proper orientation of the catalytic histidine and structural integrity of the enzyme.
J.Biol.Chem., 276, 2001
|
|
1IL1
 
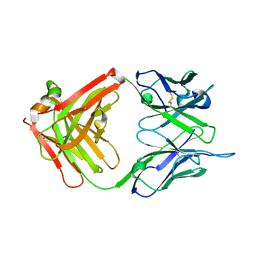 | | Crystal structure of G3-519, an anti-HIV monoclonal antibody | | Descriptor: | monoclonal antibody G3-519 (heavy chain), monoclonal antibody G3-519 (light chain) | | Authors: | Berry, M.B, Johnson, K.A, Radding, W, Fung, M, Liou, R, Phillips Jr, G.N. | | Deposit date: | 2001-05-07 | | Release date: | 2001-05-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of an anti-HIV monoclonal Fab antibody fragment specific to a gp120 C-4 region peptide.
Proteins, 45, 2001
|
|
1IL2
 
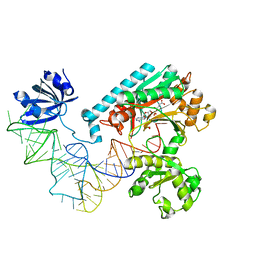 | | Crystal Structure of the E. coli Aspartyl-tRNA Synthetase:Yeast tRNAasp:aspartyl-Adenylate Complex | | Descriptor: | ASPARTYL TRANSFER RNA, ASPARTYL-ADENOSINE-5'-MONOPHOSPHATE, ASPARTYL-TRNA SYNTHETASE, ... | | Authors: | Moulinier, L, Eiler, S, Eriani, G, Gangloff, J, Thierry, J.C, Gabriel, K, McClain, W.H, Moras, D. | | Deposit date: | 2001-05-07 | | Release date: | 2001-09-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of an AspRS-tRNA(Asp) complex reveals a tRNA-dependent control mechanism.
EMBO J., 20, 2001
|
|
1IL3
 
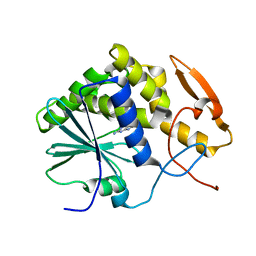 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 7-DEAZAGUANINE | | Descriptor: | 7-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL4
 
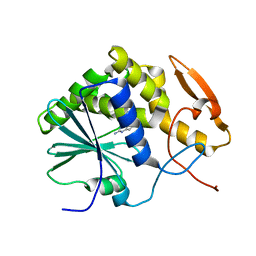 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 9-DEAZAGUANINE | | Descriptor: | 9-DEAZAGUANINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL5
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 2,5-DIAMINO-4,6-DIHYDROXYPYRIMIDINE (DDP) | | Descriptor: | 2,4-DIAMINO-4,6-DIHYDROXYPYRIMIDINE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1IL9
 
 | | STRUCTURE OF RICIN A CHAIN BOUND WITH INHIBITOR 8-METHYL-9-OXOGUANINE | | Descriptor: | 5-AMINO-2-METHYL-6H-OXAZOLO[5,4-D]PYRIMIDIN-7-ONE, RICIN A CHAIN | | Authors: | Miller, D.J, Ravikumar, K, Shen, H, Suh, J.-K, Kerwin, S.M, Robertus, J.D. | | Deposit date: | 2001-05-07 | | Release date: | 2002-01-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design and characterization of novel platforms for ricin and shiga toxin inhibition.
J.Med.Chem., 45, 2002
|
|
1ILC
 
 | | DNA Bending by an Adenine-Thymine Tract and Its Role in Gene Regulation. | | Descriptor: | 5'-D(*AP*CP*CP*GP*AP*AP*TP*TP*CP*GP*GP*T)-3' | | Authors: | Hizver, J, Rozenberg, H, Frolow, F, Rabinovich, D, Shakked, Z. | | Deposit date: | 2001-05-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | DNA bending by an adenine--thymine tract and its role in gene regulation.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1ILD
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1ILG
 
 | |
1ILH
 
 | |
1ILK
 
 | |
1ILR
 
 | |
1ILV
 
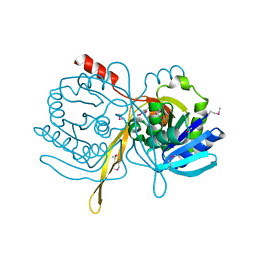 | | Crystal Structure Analysis of the TM107 | | Descriptor: | STATIONARY-PHASE SURVIVAL PROTEIN SURE HOMOLOG | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Savchenko, A, Beasley, S, Evdokimova, E, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase.
Structure, 9, 2001
|
|
1ILW
 
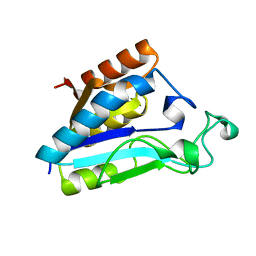 | |
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1IM0
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT PH 8.3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHSOPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
1IM2
 
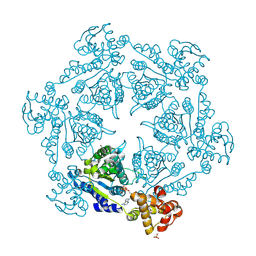 | | HslU, Haemophilus Influenzae, Selenomethionine Variant | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-DEPENDENT HSL PROTEASE ATP-BINDING SUBUNIT HSLU, SULFATE ION | | Authors: | Trame, C.B, McKay, D.B. | | Deposit date: | 2001-05-09 | | Release date: | 2001-08-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
