5ZD0
 
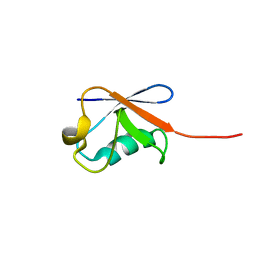 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZEV
 
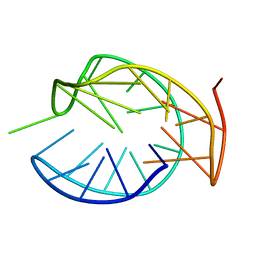 | |
5ZEW
 
 | |
5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
5ZGG
 
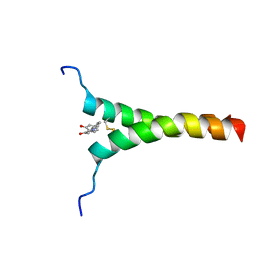 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
5ZKV
 
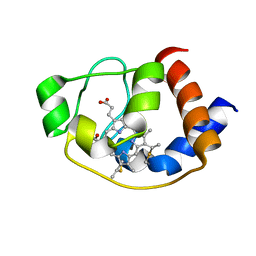 | | Solution structure of molten globule state of L94G mutant of horse cytochrome-c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Naiyer, A, Islam, A, Hassan, M.I, Sundd, M, Ahmad, F. | | Deposit date: | 2018-03-26 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of molten globule state of L94G mutant of horse cytochrome-c
To Be Published
|
|
5ZLD
 
 | |
5ZMB
 
 | |
5ZMR
 
 | | Solution Structure of the N-terminal Domain of the Yeast Rpn5 | | Descriptor: | 26S proteasome regulatory subunit RPN5 | | Authors: | Zhang, W, Zhao, C, Li, H, Hu, Y, Jin, C. | | Deposit date: | 2018-04-05 | | Release date: | 2018-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal domain of proteasome lid subunit Rpn5
Biochem. Biophys. Res. Commun., 504, 2018
|
|
5ZNU
 
 | | Structure of omega conotoxin Bu8 | | Descriptor: | Omega-conotoxin-like Bu8 | | Authors: | Liu, X, Jiang, L. | | Deposit date: | 2018-04-11 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of omega conotoxin bu8
To Be Published
|
|
5ZOR
 
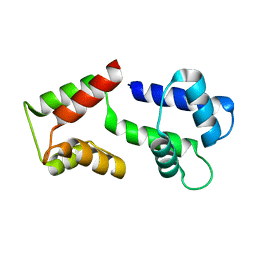 | |
5ZPV
 
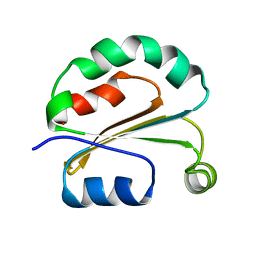 | |
5ZR0
 
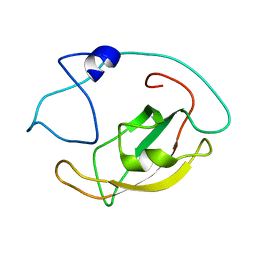 | | Solution structure of peptidyl-prolyl cis/trans isomerase domain of Trigger Factor in complex with MBP | | Descriptor: | Maltose-binding periplasmic protein,Trigger factor | | Authors: | Kawagoe, S, Nakagawa, H, Kumeta, H, Ishimori, K, Saio, T. | | Deposit date: | 2018-04-21 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into prolinecis/transisomerization of unfolded proteins catalyzed by the trigger factor chaperone.
J. Biol. Chem., 293, 2018
|
|
5ZSW
 
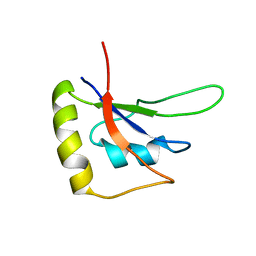 | | RBM10-RRM2 domain and its lung cancer related mutant | | Descriptor: | RNA-binding protein 10 | | Authors: | Qin, H. | | Deposit date: | 2018-04-30 | | Release date: | 2019-05-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamics, and RNA binding comparison between RBM10-RRM2 domain and its lung cancer related mutation.
To be published
|
|
5ZSY
 
 | | RBM10-RRM2 domain and its lung cancer related mutant | | Descriptor: | RNA-binding protein 10 | | Authors: | Qin, H. | | Deposit date: | 2018-04-30 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural, dynamics, and RNA binding comparison between RBM10-RRM2 domain and its lung cancer related mutation.
To be published
|
|
5ZUH
 
 | |
5ZUX
 
 | |
5ZUZ
 
 | |
5ZV6
 
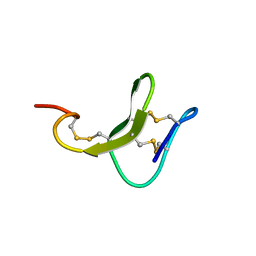 | |
5ZVF
 
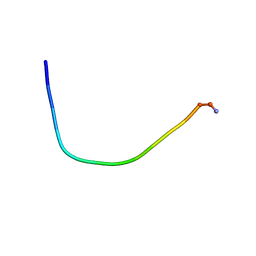 | | Structure of [T9,K7]indolicidin, a non glycosylated analogue of indolicidin | | Descriptor: | non glycosylated analogue of Indolicidin | | Authors: | Dwivedi, R, Aggarwal, P, Kaur, K.J, Bhavesh, N.S. | | Deposit date: | 2018-05-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Design of therapeutically improved analogue of the antimicrobial peptide, indolicidin, using a glycosylation strategy.
Amino Acids, 51, 2019
|
|
5ZVN
 
 | | Structure of [beta Glc-T9,K7]indolicidin, a glycosylated analogue of indolicidin | | Descriptor: | beta-D-glucopyranose, glycosylated analogue of Indolicidin | | Authors: | Dwivedi, R, Aggarwal, P, Kaur, K.J, Bhavesh, N.S. | | Deposit date: | 2018-05-11 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Design of therapeutically improved analogue of the antimicrobial peptide, indolicidin, using a glycosylation strategy.
Amino Acids, 51, 2019
|
|
5ZYX
 
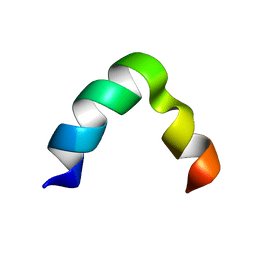 | | Solution NMR structure of K30 peptide in 10 mM dioctanoyl phosphatidylglycerol (D8PG) | | Descriptor: | ARG-TRP-LYS-ARG-HIS-ILE-SER-GLU-GLN-LEU-ARG-ARG-ARG-ASP-ARG-LEU-GLN-ARG-GLN-ALA | | Authors: | Bhunia, A, Mohid, A, Stella, L, Calligari, P. | | Deposit date: | 2018-05-28 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design, Synthesis, Antibacterial Potential, and Structural Characterization of N-Acylated Derivatives of the Human Autophagy 16 Polypeptide.
Bioconjug.Chem., 30, 2019
|
|
6A3Z
 
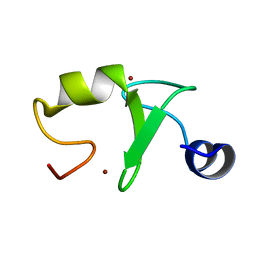 | | Zinc finger domain from the HRD1 Protein | | Descriptor: | E3 ubiquitin-protein ligase synoviolin, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2018-06-18 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structure of the zinc finger from the HRD1 protein
To Be Published
|
|
6A4C
 
 | | Solution structure of MXAN_0049 | | Descriptor: | Uncharacterized protein MXAN_0049 | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2018-06-19 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MXAN_0049
To Be Published
|
|
6A5I
 
 | | Pseudocerastes Persicus Trypsin Inhibitor | | Descriptor: | Trypsin Inhibitor | | Authors: | Amininasab, M. | | Deposit date: | 2018-06-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of PPTI, a kunitz-type protein from the venom of Pseudocerastes persicus.
PLoS ONE, 14, 2019
|
|
