2NCV
 
 | | NMR structure of RWS21 structure in LPS micelles | | Descriptor: | Heparin cofactor 2 | | Authors: | Datta, A, Bhunia, A. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Effects of N-Terminal Modifications of the Anti-Inflammatory Peptide KYE21 on Lipopolysaccharide and Membrane Interactions
To be Published
|
|
2NCW
 
 | |
2NCX
 
 | |
2NCY
 
 | | Solution structure of pseudin-2 analog (Ps-P) | | Descriptor: | Pseudin-2 | | Authors: | Jeon, D, Kim, J, Shin, A, Kim, Y. | | Deposit date: | 2016-04-18 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Optimum Balance between the Cationicity and Structural Component for Bacterial Cell Selectivity and Anti-inflammatory activities of Pseudin-2 and its Analogs
To be Published
|
|
2NCZ
 
 | |
2ND0
 
 | |
2ND1
 
 | |
2ND2
 
 | | Solution structure of the de novo mini protein gHHH_06 | | Descriptor: | De novo mini protein HHH_06 | | Authors: | Pulavarti, S.V, Eletsky, A, Bahl, C.D, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND3
 
 | | Solution structure of the de novo mini protein gEEH_04 | | Descriptor: | De novo mini protein EEH_04 | | Authors: | Pulavarti, S.V, Bahl, C.D, Gilmore, J.M, Eletsky, A, Buchko, G.W, Baker, D, Szyperski, T. | | Deposit date: | 2016-04-22 | | Release date: | 2016-09-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Accurate de novo design of hyperstable constrained peptides.
Nature, 538, 2016
|
|
2ND4
 
 | |
2ND5
 
 | | Lysine dimethylated FKBP12 | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Hattori, Y, Sebera, J, Sychrovsky, V, Furuita, K, Sugiki, T, Ohki, I, Ikegami, T, Kobayashi, N, Tanaka, Y, Fujiwara, T, Kojima, C. | | Deposit date: | 2016-05-05 | | Release date: | 2017-05-17 | | Method: | SOLUTION NMR | | Cite: | NMR Observation of Protein Surface Salt Bridges at Neutral pH
To be Published
|
|
2ND6
 
 | | Structure of DK17 in GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND7
 
 | | Structure of DK17 in POPC:POPG:Cholesterol:GM1 LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND8
 
 | | Structures of DK17 in TBLE LUVS | | Descriptor: | Cell penetrating peptide | | Authors: | Bera, S, Bhunia, A. | | Deposit date: | 2016-05-11 | | Release date: | 2017-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Elucidation of the Cell-Penetrating Penetratin Peptide in Model Membranes at the Atomic Level: Probing Hydrophobic Interactions in the Blood-Brain Barrier
Biochemistry, 55, 2016
|
|
2ND9
 
 | | Solution structure of MapZ extracellular domain first subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2NDA
 
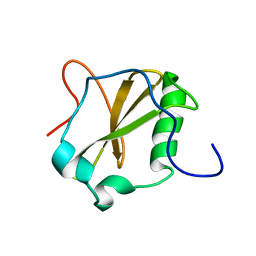 | | Solution structure of MapZ extracellular domain second subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2NDB
 
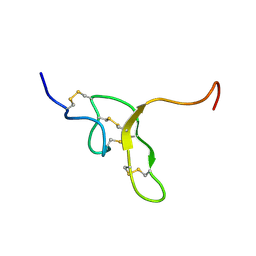 | |
2NDC
 
 | | Solution Structure of BMAP-28(1-18) | | Descriptor: | Cathelicidin-5 | | Authors: | Agadi, N, Kumar, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Function And Membrane Interaction Studies Of Two Synthetic Antimicrobial Peptides Using Solution And Solid State NMR
To be Published
|
|
2NDD
 
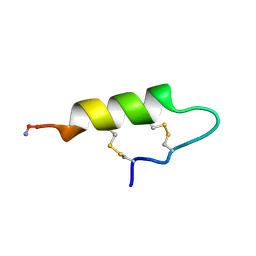 | |
2NDE
 
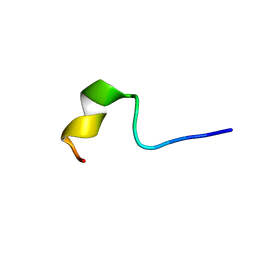 | | Solution Structure of Mutant of BMAP-28(1-18) | | Descriptor: | Cathelicidin-5 | | Authors: | Agadi, N, Kumar, A. | | Deposit date: | 2016-05-12 | | Release date: | 2017-09-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Function And Membrane Interaction Studies of Two Synthetic Peptides Using Solution And Solid State NMR
To be Published
|
|
2NDF
 
 | |
2NDG
 
 | |
2NDH
 
 | | NMR solution structure of MAL/TIRAP TIR domain (C116A) | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Lavrencic, P, Mobli, M. | | Deposit date: | 2016-05-27 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TLR adaptor MAL/TIRAP reveals an intact BB loop and supports MAL Cys91 glutathionylation for signaling.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
2NDI
 
 | | Solution structure of the toxin ISTX-I from Ixodes scapularis | | Descriptor: | Putative secreted salivary protein | | Authors: | Hu, K. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A sodium channel inhibitor ISTX-I with a novel structure provides a new hint at the evolutionary link between two toxin folds.
Sci Rep, 6, 2016
|
|
2NDJ
 
 | | Structural Basis for KCNE3 and Estrogen Modulation of the KCNQ1 Channel | | Descriptor: | Potassium voltage-gated channel subfamily E member 3 | | Authors: | Sanders, C.R, Van Horn, W.D, Kroncke, B.M, Sisco, N.J, Meiler, J, Vanoye, C.G, Song, Y, Nannemann, D.P, Welch, R.C, Kang, C, Smith, J, George, A.L. | | Deposit date: | 2016-06-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for KCNE3 modulation of potassium recycling in epithelia.
Sci Adv, 2, 2016
|
|
