2L8F
 
 | |
2L8H
 
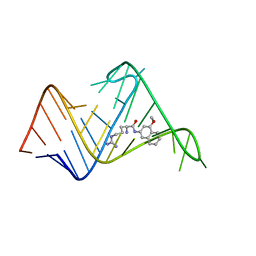 | | Chemical probe bound to HIV TAR RNA | | Descriptor: | 4-methoxynaphthalen-2-amine, ARGININE, HIV TAR RNA | | Authors: | Davidson, A, Begley, D, Lau, C, Varani, G. | | Deposit date: | 2011-01-12 | | Release date: | 2011-03-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Small-Molecule Probe Induces a Conformation in HIV TAR RNA Capable of Binding Drug-Like Fragments.
J.Mol.Biol., 410, 2011
|
|
2L8I
 
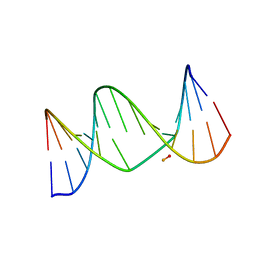 | | A biocompatible backbone modification? - Structure and dynamics of a triazole-linked DNA duplex | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*G*(2L8)P*TP*GP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*AP*AP*AP*CP*GP*TP*CP*G)-3') | | Authors: | El-Sagheer, A, Brown, T, Ernsting, N, Dehmel, L, Griesinger, C, Mugge, C. | | Deposit date: | 2011-01-13 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained.
Chemistry, 17, 2011
|
|
2L8J
 
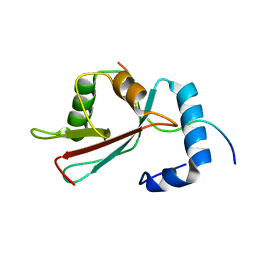 | | GABARAPL-1 NBR1-LIR complex structure | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, NBR1-LIR peptide | | Authors: | Rogov, V.V, Rozenknop, A, Rogova, N.Y, Loehr, F, Guentert, P, Dikic, I, Doetsch, V. | | Deposit date: | 2011-01-17 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1.
J.Mol.Biol., 410, 2011
|
|
2L8K
 
 | | NMR Structure of the Arterivirus nonstructural protein 7 alpha (nsp7 alpha) | | Descriptor: | Non-structural protein 7 | | Authors: | Conte, M.R, Gaudin, C, Manolaridis, I, Tucker, P.W, Kelly, G. | | Deposit date: | 2011-01-19 | | Release date: | 2011-07-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Genetic Analysis of the Arterivirus Nonstructural Protein 7{alpha}.
J.Virol., 85, 2011
|
|
2L8L
 
 | |
2L8M
 
 | | Reduced and CO-bound cytochrome P450cam (CYP101A1) | | Descriptor: | CAMPHOR, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Pochapsky, T.C, Pochapsky, S.S, Dang, M, Asciutto, E, Madura, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-02-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Experimentally Restrained Molecular Dynamics Simulations for Characterizing the Open States of Cytochrome P450(cam).
Biochemistry, 50, 2011
|
|
2L8N
 
 | |
2L8O
 
 | | Solution structure of Chr148 from Cytophaga hutchinsonii, Northeast Structural Genomics Consortium Target Chr148 | | Descriptor: | Activator of Hsp90 ATPase homologue 1-like C-terminal domain-containing protein | | Authors: | Liu, Y, Lee, D, Ciccosanti, C, Nair, L, Rost, B, Acton, T, Xiao, R, Everett, J, Montelione, G, Prestegard, J, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-21 | | Release date: | 2011-03-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Chr148 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target Chr148
To be Published
|
|
2L8P
 
 | | Solution Structure of a DNA Duplex Containing the Potent Anti-Poxvirus Agent Cidofovir | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*GP*(L8P)P*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Julien, O, Beadle, J.R, Magee, W.C, Chatterjee, S, Hostetler, K.Y, Evans, D.H, Sykes, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
J.Am.Chem.Soc., 133, 2011
|
|
2L8Q
 
 | | Solution Structure of a control DNA Duplex | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*TP*GP*CP*TP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*AP*GP*CP*AP*TP*GP*CP*G)-3') | | Authors: | Julien, O, Beadle, J.R, Magee, W.C, Chatterjee, S, Hostetler, K.Y, Evans, D.H, Sykes, B.D. | | Deposit date: | 2011-01-22 | | Release date: | 2011-02-23 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex containing the potent anti-poxvirus agent cidofovir.
J.Am.Chem.Soc., 133, 2011
|
|
2L8R
 
 | |
2L8S
 
 | |
2L8T
 
 | | Staphylococcus aureus pathogenicity island 1 protein gp6, an internal scaffold in size determination | | Descriptor: | Transposon Tn557 toxic shock syndrome toxin-1 | | Authors: | Dearborn, A.D, Spilman, M.S, Damle, P.K, Chang, J.R, Monroe, E.B, Saad, J.S, Christie, G.E, Dokland, T. | | Deposit date: | 2011-01-24 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Staphylococcus aureus Pathogenicity Island 1 Protein gp6 Functions as an Internal Scaffold during Capsid Size Determination.
J.Mol.Biol., 412, 2011
|
|
2L8U
 
 | |
2L8V
 
 | | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC (20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics Consortium Target TeR219A | | Descriptor: | Phycobilisome 32.1 kDa linker polypeptide, phycocyanin-associated, rod | | Authors: | Ramelot, T.A, Yang, Y, Cort, J.R, Lee, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-01-26 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the phycobilisome linker polypeptide domain of CpcC
(20-153) from Thermosynechococcus elongatus, Northeast Structural Genomics
Consortium Target TeR219A
To be Published
|
|
2L8W
 
 | |
2L8X
 
 | |
2L8Y
 
 | | Solution structure of the E. coli outer membrane protein RcsF (periplasmatic domain) | | Descriptor: | Protein rcsF | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Lohr, F, Doetsch, V. | | Deposit date: | 2011-01-27 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF.
J.Biol.Chem., 286, 2011
|
|
2L90
 
 | | Solution structure of murine myristoylated msrA | | Descriptor: | MYRISTIC ACID, Peptide methionine sulfoxide reductase | | Authors: | Gruschus, J.M, Lim, J, Piszczek, G, Levine, R.L, Tjandra, N. | | Deposit date: | 2011-01-27 | | Release date: | 2012-01-11 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Characterization and solution structure of mouse myristoylated methionine sulfoxide reductase A.
J.Biol.Chem., 287, 2012
|
|
2L91
 
 | |
2L92
 
 | |
2L93
 
 | |
2L94
 
 | | Structure of the HIV-1 frameshift site RNA bound to a small molecule inhibitor of viral replication | | Descriptor: | N'-{(Z)-amino[4-(amino{[3-(dimethylammonio)propyl]iminio}methyl)phenyl]methylidene}-N,N-dimethylpropane-1,3-diaminium, RNA_(45-MER) | | Authors: | Marcheschi, R.J, Tonelli, M, Kumar, A, Butcher, S.E. | | Deposit date: | 2011-01-29 | | Release date: | 2011-06-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 Frameshift Site RNA Bound to a Small Molecule Inhibitor of Viral Replication.
Acs Chem.Biol., 6, 2011
|
|
2L95
 
 | | Solution Structure of Cytotoxic T-Lymphocyte Antigent-2(Ctla protein), Crammer at pH 6.0 | | Descriptor: | Crammer | | Authors: | Tseng, T.S, Cheng, C.S, Liu, Y.N, Lyu, P.C. | | Deposit date: | 2011-01-31 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A molten globule-to-ordered structure transition of Drosophila melanogaster crammer is required for its ability to inhibit cathepsin.
Biochem.J., 442, 2012
|
|
