2ARI
 
 | | Solution structure of micelle-bound fusion domain of HIV-1 gp41 | | Descriptor: | Envelope polyprotein GP160 | | Authors: | Jaroniec, C.P, Kaufman, J.D, Stahl, S.J, Viard, M, Blumenthal, R, Wingfield, P.T, Bax, A. | | Deposit date: | 2005-08-19 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of Micelle-Associated Human Immunodeficiency Virus gp41 Fusion Domain.
Biochemistry, 44, 2005
|
|
2ASE
 
 | |
2ASQ
 
 | | Solution Structure of SUMO-1 in Complex with a SUMO-binding Motif (SBM) | | Descriptor: | Protein inhibitor of activated STAT2, Small ubiquitin-related modifier 1 | | Authors: | Song, J, Zhang, Z, Hu, W, Chen, Y. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small Ubiquitin-like Modifier (SUMO) Recognition of a SUMO Binding Motif: A reversal of the bound orientation
J.Biol.Chem., 280, 2005
|
|
2ASY
 
 | | Solution Structure of ydhR protein from Escherichia coli | | Descriptor: | Protein ydhR precursor | | Authors: | Revington, M, Semesi, A, Yee, A, Shaw, G.S, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2005-08-24 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Escherichia coli protein ydhR: A putative mono-oxygenase.
Protein Sci., 14, 2005
|
|
2ATG
 
 | | NMR structure of Retrocyclin-2 in SDS | | Descriptor: | Retrocyclin-2 | | Authors: | Daly, N.L, Chen, Y.K, Rosengren, K.J, Marx, U.C, Phillips, M.L, Waring, A.J, Wang, W, Lehrer, R.I, Craik, D.J. | | Deposit date: | 2005-08-24 | | Release date: | 2005-09-06 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Retrocyclin-2: structural analysis of a potent anti-HIV theta-defensin
Biochemistry, 46, 2007
|
|
2AU4
 
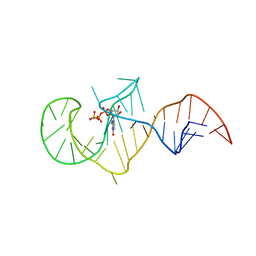 | | Class I GTP aptamer | | Descriptor: | Class I RNA aptamer to GTP, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Carothers, J.M, Davis, J.H, Chou, J.J, Szostak, J.W. | | Deposit date: | 2005-08-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an informationally complex high-affinity RNA aptamer to GTP.
Rna, 12, 2006
|
|
2AUV
 
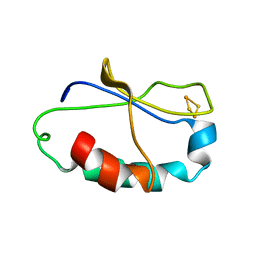 | | Solution Structure of HndAc : A Thioredoxin-like [2Fe-2S] Ferredoxin Involved in the NADP-reducing Hydrogenase Complex | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, potential NAD-reducing hydrogenase subunit | | Authors: | Nouailler, M, Morelli, X, Bornet, O, Chetrit, B, Dermoun, Z, Guerlesquin, F. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of HndAc: a thioredoxin-like domain involved in the NADP-reducing hydrogenase complex
Protein Sci., 15, 2006
|
|
2AVA
 
 | |
2AVG
 
 | |
2AWQ
 
 | |
2AWT
 
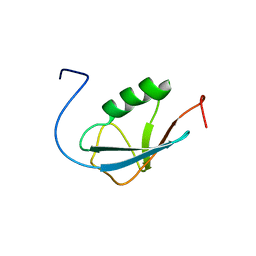 | | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2) | | Descriptor: | Small ubiquitin-related modifier 2 | | Authors: | Chang, C.K, Wang, Y.H, Chung, T.L, Chang, C.F, Li, S.S.L, Huang, T.H. | | Deposit date: | 2005-09-02 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human Small Ubiquitin-Like Modifier Protein Isoform 2 (SUMO-2)
To be Published
|
|
2AWV
 
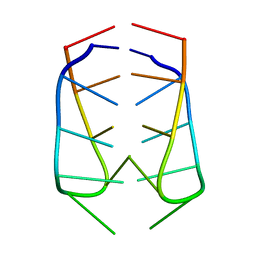 | | NMR Structural Analysis of the dimer of 5MCCTCATCC | | Descriptor: | 5'-D(*(MCY)P*CP*TP*CP*AP*CP*TP*CP*C)-3' | | Authors: | Canalia, M, Leroy, J.-L. | | Deposit date: | 2005-09-02 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure, internal motions and association-dissociation kinetics of the i-motif dimer of d(5mCCTCACTCC).
Nucleic Acids Res., 33, 2005
|
|
2AX5
 
 | | Solution Structure of Urm1 from Saccharomyces Cerevisiae | | Descriptor: | Hypothetical 11.0 kDa protein in FAA3-MAS3 intergenic region | | Authors: | Xu, J, Huang, H, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2005-09-03 | | Release date: | 2006-06-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Urm1 and its implications for the origin of protein modifiers.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2AXD
 
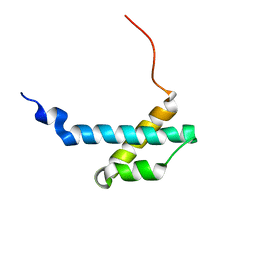 | | solution structure of the theta subunit of escherichia coli DNA polymerase III in complex with the epsilon subunit | | Descriptor: | DNA polymerase III, theta subunit | | Authors: | Keniry, M.A, Park, A.Y, Owen, E.A, Hamdan, S.M, Pintacuda, G, Otting, G, Dixon, N.E. | | Deposit date: | 2005-09-05 | | Release date: | 2006-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the theta subunit of Escherichia coli DNA polymerase III in complex with the epsilon subunit
J.Bacteriol., 188, 2006
|
|
2AXK
 
 | | Solution structure of discrepin, a scorpion venom toxin blocking K+ channels. | | Descriptor: | discrepin | | Authors: | Prochnicka-Chalufour, A, Corzo, G, Satake, H, Martin-Eauclaire, M.-F, Murgia, A.R, Prestipino, G, D'Suze, G, Possani, L.D, Delepierre, M. | | Deposit date: | 2005-09-05 | | Release date: | 2006-06-20 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of discrepin, a new K+-channel blocking peptide from the alpha-KTx15 subfamily.
Biochemistry, 45, 2006
|
|
2AXL
 
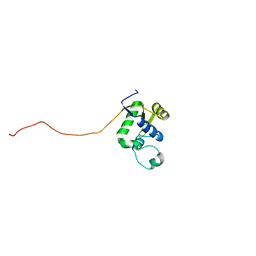 | | Solution structure of a multifunctional DNA- and protein-binding domain of human Werner syndrome protein | | Descriptor: | Werner syndrome | | Authors: | Hu, J.-S, Feng, H, Zeng, W, Lin, G.-X, Xi, X.G. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a multifunctional DNA- and protein-binding motif of human Werner syndrome protein.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AYA
 
 | |
2AYJ
 
 | | Solution structure of 50S ribosomal protein L40e from Sulfolobus solfataricus | | Descriptor: | 50S ribosomal protein L40e, ZINC ION | | Authors: | Wu, B, Yee, A, Lukin, J, Lemak, A, Semesi, A, Ramelot, T, Kennedy, M, Edward, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-07 | | Release date: | 2006-08-22 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of ribosomal protein L40E, a unique C4 zinc finger protein encoded by archaeon Sulfolobus solfataricus
Protein Sci., 17, 2008
|
|
2AYM
 
 | |
2AYX
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-949) containing linker region and phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYY
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 700-816) containing linker region | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AYZ
 
 | | Solution structure of the E.coli RcsC C-terminus (residues 817-949) containing phosphoreceiver domain | | Descriptor: | Sensor kinase protein rcsC | | Authors: | Rogov, V.V, Rogova, N.Y, Bernhard, F, Koglin, A, Lohr, F, Dotsch, V. | | Deposit date: | 2005-09-09 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A New Structural Domain in the Escherichia coli RcsC Hybrid Sensor Kinase Connects Histidine Kinase and Phosphoreceiver Domains
J.Mol.Biol., 364, 2006
|
|
2AZH
 
 | | Solution structure of iron-sulfur cluster assembly protein SUFU from Bacillus subtilis, with zinc bound at the active site. Northeast Structural Genomics Consortium target SR17 | | Descriptor: | SufU, ZINC ION | | Authors: | Kornhaber, G.J, Swapna, G.V.T, Ramelot, T.A, Cort, J.R, Aramini, J.M, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-09-10 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Zn-Ligated Fe-S Cluster Assembly Scaffold Protein SufU From Bacillus subtilis
To be published
|
|
2AZS
 
 | | NMR structure of the N-terminal SH3 domain of Drk (calculated without NOE restraints) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2AZV
 
 | | Solution structure of the T22G mutant of N-terminal SH3 domain of DRK (calculated without NOEs) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
