1R4T
 
 | | Solution structure of exoenzyme S | | Descriptor: | exoenzyme S | | Authors: | Langdon, G.M, Leitner, D, Labudde, D, Kuhne, R, Schmieder, P, Aktories, K, Oschkinat, H.O, Schmidt, G. | | Deposit date: | 2003-10-08 | | Release date: | 2005-04-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal GTPase activating domain of Pseudomonas aeruginosa exoenzyme S
To be Published
|
|
1R4Y
 
 | | SOLUTION STRUCTURE OF THE DELETION MUTANT DELTA(7-22) OF THE CYTOTOXIC RIBONUCLEASE ALPHA-SARCIN | | Descriptor: | Ribonuclease alpha-sarcin | | Authors: | Garcia-Mayoral, M.F, Garcia-Ortega, L, Lillo, M.P, Santoro, J, Martinez Del Pozo, A, Gavilanes, J.G, Rico, M, Bruix, M. | | Deposit date: | 2003-10-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the noncytotoxic {alpha}-sarcin mutant {Delta}(7-22): The importance of the native conformation of peripheral loops for activity.
Protein Sci., 13, 2004
|
|
1R57
 
 | | NMR Solution Structure of a GCN5-like putative N-acetyltransferase from Staphylococcus aureus. Northeast Structural Genomics Consortium Target ZR31 | | Descriptor: | conserved hypothetical protein | | Authors: | Cort, J.R, Acton, T.B, Ma, L, Xiao, R.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-09 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
1R5S
 
 | | Connexin 43 Carboxyl Terminal Domain | | Descriptor: | Gap junction alpha-1 protein | | Authors: | Sorgen, P.L, Duffy, H.S, Mario, D, Sahoo, P, Coombs, W, Delmar, M, Spray, D.C. | | Deposit date: | 2003-10-13 | | Release date: | 2004-10-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural changes in the carboxyl terminus of the gap junction protein connexin43 indicates signaling between binding domains for c-Src and zonula occludens-1
J.Biol.Chem., 279, 2004
|
|
1R63
 
 | | STRUCTURAL ROLE OF A BURIED SALT BRIDGE IN THE 434 REPRESSOR DNA-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | REPRESSOR PROTEIN FROM BACTERIOPHAGE 434 | | Authors: | Pervushin, K.V, Billeter, M, Siegal, G, Wuthrich, K. | | Deposit date: | 1996-11-08 | | Release date: | 1997-06-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural role of a buried salt bridge in the 434 repressor DNA-binding domain.
J.Mol.Biol., 264, 1996
|
|
1R6H
 
 | | Solution Structure of human PRL-3 | | Descriptor: | protein tyrosine phosphatase type IVA, member 3 isoform 1 | | Authors: | Kozlov, G, Gehring, K, Ekiel, I. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into Molecular Function of the Metastasis-associated Phosphatase PRL-3.
J.Biol.Chem., 279, 2004
|
|
1R6P
 
 | | NMR structure of the N-terminal domain of trout cardiac troponin C at 7 C | | Descriptor: | CALCIUM ION, troponin C | | Authors: | Blumenschein, T.M, Gillis, T.E, Tibbits, G.F, Sykes, B.D. | | Deposit date: | 2003-10-15 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Effect of temperature on the structure of trout troponin C
Biochemistry, 43, 2004
|
|
1R6R
 
 | | Solution Structure of Dengue Virus Capsid Protein Reveals a New Fold | | Descriptor: | Genome polyprotein | | Authors: | Ma, L, Jones, C.T, Groesch, T.D, Kuhn, R.J, Post, C.B. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dengue virus capsid protein reveals another fold
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1R73
 
 | | Solution Structure of TM1492, the L29 ribosomal protein from Thermotoga maritima | | Descriptor: | 50S ribosomal protein L29 | | Authors: | Peti, W, Etezady-Esfarjani, T, Herrmann, T, Klock, H.E, Lesley, S.A, Wuethrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2003-10-17 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR for structural proteomics of Thermotoga maritima: Screening and structure determination
J.STRUCT.FUNCT.GENOM., 5, 2004
|
|
1R7C
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7F
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 43 structures. Sample in 100mM SDS) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1R7W
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R7Z
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1R84
 
 | | NMR structure of the 13-cis-15-syn retinal in dark_adapted bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Patzelt, H, Simon, B, Ter Laak, A, Kessler, B, Kuhne, R, Schmieder, P, Oesterhaelt, D, Oschkinat, H. | | Deposit date: | 2003-10-23 | | Release date: | 2003-11-11 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The structures of the active center in dark-adapted bacteriorhodopsin by solution-state NMR spectroscopy
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1R8P
 
 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1R8T
 
 | | Solution structures of high affinity miniprotein ligands to Streptavidin | | Descriptor: | MP1 | | Authors: | Luo, J, Mukherjee, M, Fan, X, Yang, H, Liu, D, Khan, R, White, M, Fox, R.O. | | Deposit date: | 2003-10-28 | | Release date: | 2005-02-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structure-based design of high affinity miniprotein ligands
To be Published
|
|
1R8U
 
 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
1R9I
 
 | | NMR Solution Structure of PIIIA toxin, NMR, 20 structures | | Descriptor: | Mu-conotoxin PIIIA | | Authors: | Nielsen, K.J, Watson, M, Adams, D.J, Hammarstrom, A.K, Gage, P.W, Hill, J.M, Craik, D.J, Thomas, L, Adams, D, Alewood, P.F, Lewis, R.J. | | Deposit date: | 2003-10-30 | | Release date: | 2003-11-18 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of mu-conotoxin PIIIA, a preferential inhibitor of persistent tetrodotoxin-sensitive sodium channels
J.Biol.Chem., 277, 2002
|
|
1R9P
 
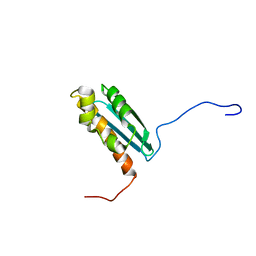 | | Solution NMR Structure Of The Haemophilus Influenzae Iron-Sulfur Cluster Assembly Protein U (IscU) with Zinc Bound at the Active Site. Northeast Structural Genomics Consortium Target IR24. | | Descriptor: | NifU-like protein, ZINC ION | | Authors: | Ramelot, T.A, Cort, J.R, Xiao, R, Shastry, R, Acton, T.B, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-10-30 | | Release date: | 2004-11-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of the iron-sulfur cluster assembly protein U (IscU) with zinc bound at the active site.
J.Mol.Biol., 344, 2004
|
|
1R9U
 
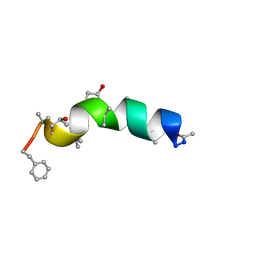 | | Refined structure of peptaibol zervamicin IIB in methanol solution from trans-hydrogen bond J couplings | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balashova, T.A, Yakimenko, Z.A, Ovchinnikova, T.V, Arseniev, A.S. | | Deposit date: | 2003-10-31 | | Release date: | 2004-11-09 | | Last modified: | 2018-10-10 | | Method: | SOLUTION NMR | | Cite: | Biosynthetic Uniform 13C,15N-Labelling of Zervamicin Iib. Complete 13C and 15N NMR Assignment.
J.Pept.Sci., 9, 2003
|
|
1R9V
 
 | |
1RCH
 
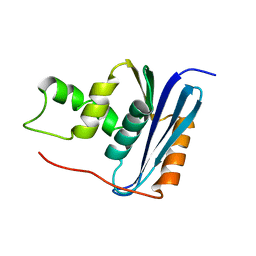 | | SOLUTION NMR STRUCTURE OF RIBONUCLEASE HI FROM ESCHERICHIA COLI, 8 STRUCTURES | | Descriptor: | RIBONUCLEASE HI | | Authors: | Yamazaki, T, Fujiwara, M, Kato, T, Yamasaki, K, Nagayama, K. | | Deposit date: | 1995-06-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ribonuclease Hi from Escherichia Coli
Biol.Pharm.Bull., 23, 2000
|
|
