1KG1
 
 | | NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis | | Descriptor: | Necrosis Inducing Protein 1 | | Authors: | Van't Slot, K.A, Van den Burg, H.A, Kloks, C.P, Hilbers, C.W, Knogge, W, Papavoine, C.H. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Plant Disease Resistance-triggering Protein NIP1 from the Fungus Rhynchosporium secalis Shows a Novel beta-Sheet Fold.
J.Biol.Chem., 278, 2003
|
|
1KGM
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-11-28 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
1KHM
 
 | | C-TERMINAL KH DOMAIN OF HNRNP K (KH3) | | Descriptor: | PROTEIN (HNRNP K) | | Authors: | Baber, J, Libutti, D, Levens, D, Tjandra, N. | | Deposit date: | 1999-01-07 | | Release date: | 2000-01-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | High precision solution structure of the C-terminal KH domain of heterogeneous nuclear ribonucleoprotein K, a c-myc transcription factor.
J.Mol.Biol., 289, 1999
|
|
1KIK
 
 | | SH3 Domain of Lymphocyte Specific Kinase (LCK) | | Descriptor: | PROTO-ONCOGENE TYROSINE-PROTEIN KINASE LCK | | Authors: | Briese, L, Willbold, D. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure determination of human Lck unique and SH3 domains by nuclear magnetic resonance spectroscopy.
Bmc Struct.Biol., 3, 2003
|
|
1KIO
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGCI[L30R, K31M] | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-12-03 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
1KIS
 
 | |
1KJ0
 
 | | SOLUTION STRUCTURE OF THE SMALL SERINE PROTEASE INHIBITOR SGTI | | Descriptor: | SERINE PROTEASE INHIBITOR I | | Authors: | Gaspari, Z, Patthy, A, Graf, L, Perczel, A. | | Deposit date: | 2001-12-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Comparative structure analysis of proteinase inhibitors from the desert locust, Schistocerca gregaria.
Eur.J.Biochem., 269, 2002
|
|
1KJS
 
 | | NMR SOLUTION STRUCTURE OF C5A AT PH 5.2, 303K, 20 STRUCTURES | | Descriptor: | C5A | | Authors: | Zhang, X, Boyar, W, Toth, M, Wennogle, L, Gonnella, N.C. | | Deposit date: | 1997-01-09 | | Release date: | 1997-05-15 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structural definition of the C5a C terminus by two-dimensional nuclear magnetic resonance spectroscopy.
Proteins, 28, 1997
|
|
1KKA
 
 | |
1KKD
 
 | | Solution structure of the calmodulin binding domain (CaMBD) of small conductance Ca2+-activated potassium channels (SK2) | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Wissmann, R, Bildl, W, Neumann, H, Rivard, A.F, Kloecker, N, Weitz, D, Schulte, U, Adelman, J.P, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin.
J.Biol.Chem., 277, 2002
|
|
1KKG
 
 | | NMR Structure of Ribosome-Binding Factor A (RbfA) | | Descriptor: | ribosome-binding factor A | | Authors: | Huang, Y.J, Swapna, G.V.T, Rajan, P.K, Ke, H, Xia, B, Shukla, K, Inouye, M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2001-12-07 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Ribosome-binding Factor A (RbfA), A Cold-shock
Adaptation Protein from Escherichia coli
J.Mol.Biol., 327, 2003
|
|
1KKV
 
 | |
1KKW
 
 | |
1KLC
 
 | | SOLUTION STRUCTURE OF TGF-B1, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | TRANSFORMING GROWTH FACTOR-BETA 1 | | Authors: | Hinck, A.P, Archer, S.J, Qian, S.W, Roberts, A.B, Sporn, M.B, Weatherbee, J.A, Tsang, M.L.-S, Lucas, R, Zhang, B.-L, Wenker, J, Torchia, D.A. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Transforming growth factor beta 1: three-dimensional structure in solution and comparison with the X-ray structure of transforming growth factor beta 2.
Biochemistry, 35, 1996
|
|
1KLQ
 
 | | The Mad2 Spindle Checkpoint Protein Undergoes Similar Major Conformational Changes upon Binding to Either Mad1 or Cdc20 | | Descriptor: | MITOTIC SPINDLE ASSEMBLY CHECKPOINT PROTEIN MAD2A, Mad2-binding peptide | | Authors: | Luo, X, Tang, Z, Rizo, J, Yu, H. | | Deposit date: | 2001-12-12 | | Release date: | 2002-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20.
Mol.Cell, 9, 2002
|
|
1KLR
 
 | | NMR Structure of the ZFY-6T[Y10F] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1KLS
 
 | | NMR Structure of the ZFY-6T[Y10L] Zinc Finger | | Descriptor: | ZINC FINGER Y-CHROMOSOMAL PROTEIN, ZINC ION | | Authors: | Lachenmann, M.J, Ladbury, J.E, Phillips, N.B, Narayana, N, Qian, X, Weiss, M.A. | | Deposit date: | 2001-12-12 | | Release date: | 2002-03-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The hidden thermodynamics of a zinc finger.
J.Mol.Biol., 316, 2002
|
|
1KMA
 
 | | NMR Structure of the Domain-I of the Kazal-type Thrombin Inhibitor Dipetalin | | Descriptor: | DIPETALIN | | Authors: | Schlott, B, Wohnert, J, Icke, C, Hartmann, M, Ramachandran, R, Guhrs, K.-H, Glusa, E, Flemming, J, Gorlach, M, Grosse, F, Ohlenschlager, O. | | Deposit date: | 2001-12-14 | | Release date: | 2002-05-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Interaction of Kazal-type inhibitor domains with serine proteinases: biochemical and structural studies.
J.Mol.Biol., 318, 2002
|
|
1KMD
 
 | | SOLUTION STRUCTURE OF THE VAM7P PX DOMAIN | | Descriptor: | Vacuolar morphogenesis protein VAM7 | | Authors: | Lu, J, Garcia, J, Dulubova, I, Sudhof, T.C, Rizo, J. | | Deposit date: | 2001-12-14 | | Release date: | 2002-06-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Vam7p PX domain.
Biochemistry, 41, 2002
|
|
1KMF
 
 | | NMR STRUCTURE OF HUMAN INSULIN MUTANT ILE-A2-ALLO-ILE, HIS-B10-ASP, PRO-B28-LYS, LYS-B29-PRO, 15 STRUCTURES | | Descriptor: | Insulin | | Authors: | Xu, B, Hua, Q.X, Nakagawa, S.H, Jia, W, Chu, Y.C, Katsoyannis, P.G, Weiss, M.A. | | Deposit date: | 2001-12-14 | | Release date: | 2002-01-09 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Chiral mutagenesis of insulin's hidden receptor-binding surface: structure of an allo-isoleucine(A2) analogue.
J.Mol.Biol., 316, 2002
|
|
1KMR
 
 | |
1KN7
 
 | | Solution structure of the tandem inactivation domain (residues 1-75) of potassium channel RCK4 (Kv1.4) | | Descriptor: | VOLTAGE-GATED POTASSIUM CHANNEL PROTEIN KV1.4 | | Authors: | Wissmann, R, Bildl, W, Oliver, D, Beyermann, M, Kalbitzer, H.R, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-18 | | Release date: | 2003-05-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Function of the "Tandem Inactivation Domain" of the Neuronal A-type
Potassium Channel Kv1.4
J.Biol.Chem., 278, 2003
|
|
1KOS
 
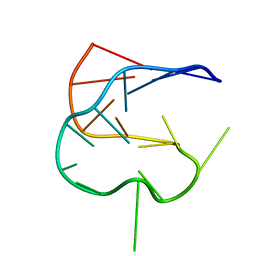 | | SOLUTION NMR STRUCTURE OF AN ANALOG OF THE YEAST TRNA PHE T STEM LOOP CONTAINING RIBOTHYMIDINE AT ITS NATURALLY OCCURRING POSITION | | Descriptor: | 5'-R(*CP*UP*GP*UP*GP*(5MU)P*UP*CP*GP*AP*UP*(CH)P*CP*AP*CP*AP*G)- 3' | | Authors: | Koshlap, K.M, Guenther, R, Sochacka, E, Malkiewicz, A, Agris, P.F. | | Deposit date: | 1999-05-03 | | Release date: | 1999-10-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | A distinctive RNA fold: the solution structure of an analogue of the yeast tRNAPhe T Psi C domain.
Biochemistry, 38, 1999
|
|
1KOT
 
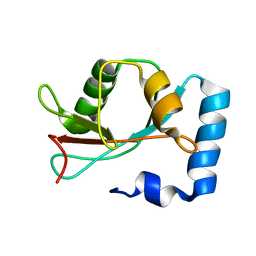 | |
1KOY
 
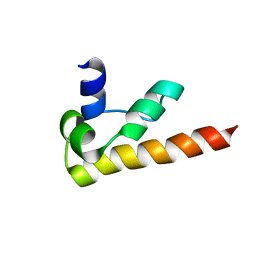 | | NMR structure of DFF-C domain | | Descriptor: | DNA fragmentation factor alpha subunit | | Authors: | Fukushima, K, Kikuchi, J, Koshiba, S, Kigawa, T, Kuroda, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-12-25 | | Release date: | 2002-09-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DFF-C domain of DFF45/ICAD. A structural basis for the regulation of apoptotic DNA fragmentation.
J.Mol.Biol., 321, 2002
|
|
