6SX6
 
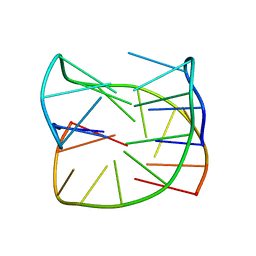 | | Guanine-rich oligonucleotide with 5'-GC end form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pairs | | Descriptor: | GCn | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-25 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
6SY2
 
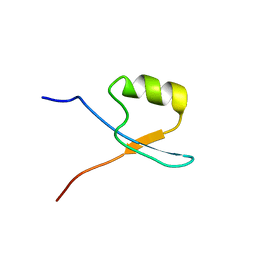 | |
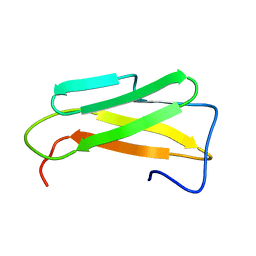
6SYK
 
 | | Guanine-rich oligonucleotide with 5'- and 3'-GC ends form G-quadruplex with A(GGGG)A hexad, GCGC- and G-quartets and two symmetric GG and AA base pair | | Descriptor: | GCnCG | | Authors: | Pavc, D, Wang, B, Spindler, L, Drevensek-Olenik, I, Plavec, J, Sket, P. | | Deposit date: | 2019-09-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | GC ends control topology of DNA G-quadruplexes and their cation-dependent assembly.
Nucleic Acids Res., 48, 2020
|
|
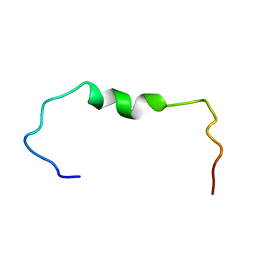
6SZC
 
 | |
6SZF
 
 | | Solution structure of the amyloid beta-peptide (1-42) | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Grimaldi, M, Santoro, A, Stillitano, I, Buonocore, M, D'Ursi, A.M. | | Deposit date: | 2019-10-02 | | Release date: | 2020-10-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Exploring the Early Stages of the Amyloid A beta (1-42) Peptide Aggregation Process: An NMR Study.
Pharmaceuticals, 14, 2021
|
|
6T2G
 
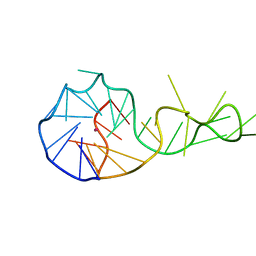 | |
6T33
 
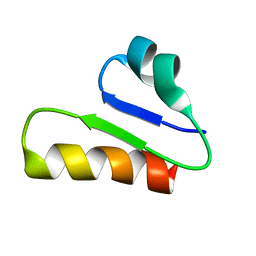 | | The unusual structure of Ruminococcin C1 antimicrobial peptide confers activity against clinical pathogens | | Descriptor: | Ruminococcin C | | Authors: | Chiumento, S, Roblin, C, Bornet, O, Nouailler, M, Muller, C, Basset, C, Kieffer-Jaquinod, S, Coute, Y, Torelli, S, Le Pape, L, Shunemann, V, Jeannot, K, Nicoletti, C, Iranzo, O, Maresca, M, Giardina, T, Fons, M, Devillard, E, Perrier, J, Atta, M, Guerlesquin, F, Lafond, M, Duarte, V. | | Deposit date: | 2019-10-10 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The unusual structure of Ruminococcin C1 antimicrobial peptide confers clinical properties.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6T3I
 
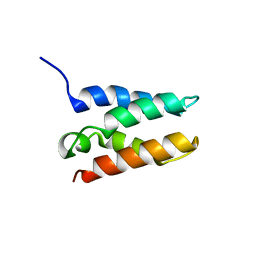 | | Solution structure of the HRP2 IBD | | Descriptor: | Hepatoma-derived growth factor-related protein 2 | | Authors: | Veverka, V. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Unlike Its Paralog LEDGF/p75, HRP-2 Is Dispensable for MLL-R Leukemogenesis but Important for Leukemic Cell Survival.
Cells, 10, 2021
|
|
6T51
 
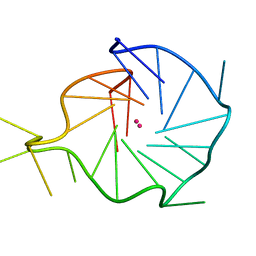 | |
6TAZ
 
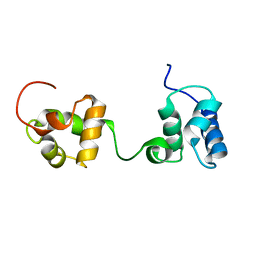 | | Timeless couples G quadruplex detection with processing by DDX11 during DNA replication | | Descriptor: | Protein timeless homolog | | Authors: | Lerner Koch, L, Holzer, S, Kilkenny, M.L, Murat, P, Svikovic, S, Schiavone, D, Bittleston, A, Maman, J.D, Branzei, D, Stott, K, Pellegrini, L, Sale, E.J. | | Deposit date: | 2019-10-31 | | Release date: | 2020-07-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication.
Embo J., 39, 2020
|
|
6TC8
 
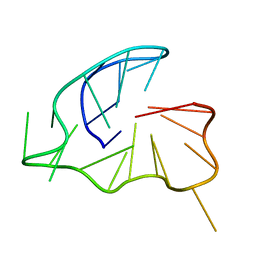 | |
6TCG
 
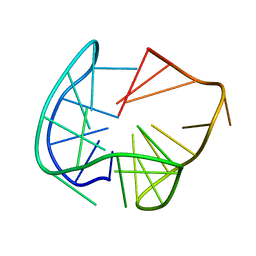 | |
6TDD
 
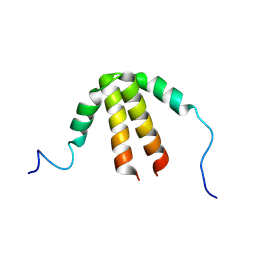 | | Bam_5924 docking domain | | Descriptor: | Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDM
 
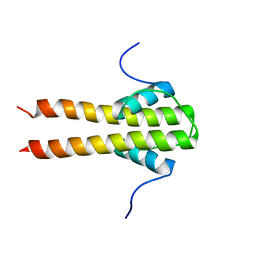 | | Bam_5920cDD 5919nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TDN
 
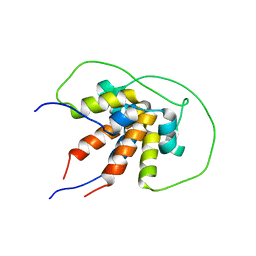 | | Bam_5925cDD 5924nDD docking domains | | Descriptor: | Beta-ketoacyl synthase,Beta-ketoacyl synthase | | Authors: | Risser, F, Chagot, B. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Towards improved understanding of intersubunit interactions in modular polyketide biosynthesis: Docking in the enacyloxin IIa polyketide synthase.
J.Struct.Biol., 212, 2020
|
|
6TEY
 
 | |
6TF4
 
 | |
6TG5
 
 | |
6TH8
 
 | |
6THI
 
 | | Solution structure of MeuNaTxalpha-1 toxin from Mesobuthus Eupeus | | Descriptor: | Sodium channel neurotoxin MeuNaTxalpha-1 | | Authors: | Mineev, K.S, Kuzmenkov, A.I, Khusainov, G.A, Arseniev, A.S, Vassilevski, A.A. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of MeuNaTx alpha-1 toxin from scorpion venom highlights the importance of the nest motif.
Proteins, 2021
|
|
6TI5
 
 | | A New Structural Model of Abeta(1-40) Fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Bertini, I, Gonnelli, L, Luchinat, C, Mao, J, Nesi, A. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI6
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TI7
 
 | | Mixing Abeta(1-40) and Abeta(1-42) peptides generates unique amyloid fibrils | | Descriptor: | Amyloid-beta precursor protein | | Authors: | Cerofolini, L, Ravera, E, Bologna, S, Wiglenda, T, Boddrich, A, Purfurst, B, Benilova, A, Korsak, M, Gallo, G, Rizzo, D, Gonnelli, L, Fragai, M, De Strooper, B, Wanker, E.E, Luchinat, C. | | Deposit date: | 2019-11-21 | | Release date: | 2020-07-22 | | Last modified: | 2024-06-19 | | Method: | SOLID-STATE NMR | | Cite: | Mixing A beta (1-40) and A beta (1-42) peptides generates unique amyloid fibrils.
Chem.Commun.(Camb.), 56, 2020
|
|
6TIQ
 
 | |
6TIR
 
 | |
