6KRA
 
 | |
6KVB
 
 | | Structure of an intra-locked G-quadruplex | | Descriptor: | DNA (28-mer) | | Authors: | Maity, A, Winnerdy, F.R, Chang, W.D, Chen, G, Phan, A.T. | | Deposit date: | 2019-09-03 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Intra-locked G-quadruplex structures formed by irregular DNA G-rich motifs.
Nucleic Acids Res., 48, 2020
|
|
6KVG
 
 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
6KXZ
 
 | |
6L0L
 
 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
6L6V
 
 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
6L7K
 
 | | solution structure of hIFABP V60C/Y70C variant. | | Descriptor: | Fatty acid-binding protein, intestinal | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ligand Entry into Fatty Acid Binding Protein via Local Unfolding Instead of Gap Widening.
Biophys.J., 118, 2020
|
|
6L7Z
 
 | | Solution NMR structure of the N-terminal immunoglobulin variable domain of BTNL2 | | Descriptor: | Butyrophilin-like protein 2 | | Authors: | Basak, A.J, Lee, W, Samanta, D, De, S. | | Deposit date: | 2019-11-03 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Structural Insights into N-terminal IgV Domain of BTNL2, a T Cell Inhibitory Molecule, Suggests a Non-canonical Binding Interface for Its Putative Receptors.
J.Mol.Biol., 432, 2020
|
|
6L87
 
 | |
6L8M
 
 | | WNT DNA promoter mutant G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
6L8R
 
 | | membrane-bound PD-L1-CD | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Maorong, W, Cao, Y, Bin, W, Bo, O. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain.
Nat Commun, 12, 2021
|
|
6L8V
 
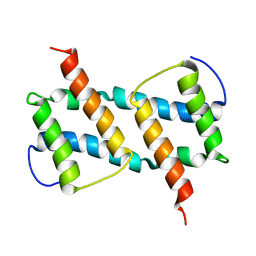 | | membrane-bound Bax helix2-helix5 domain | | Descriptor: | Apoptosis regulator BAX | | Authors: | OuYang, B, Lv, F. | | Deposit date: | 2019-11-07 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | An amphipathic Bax core dimer forms part of the apoptotic pore wall in the mitochondrial membrane.
Embo J., 40, 2021
|
|
6L92
 
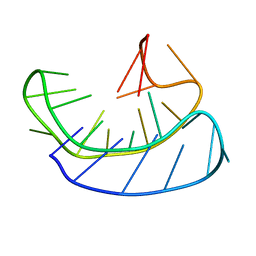 | | A basket type G-quadruplex in WNT DNA promoter | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*CP*AP*CP*CP*GP*GP*GP*CP*AP*GP*TP*GP*GP*GP*CP*GP*GP*G)-3') | | Authors: | Wang, Z.F, Li, M.H, Chu, I.T, Winnerdy, F.R, Phan, A.T, Chang, T.C. | | Deposit date: | 2019-11-08 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cytosine epigenetic modification modulates the formation of an unprecedented G4 structure in the WNT1 promoter.
Nucleic Acids Res., 48, 2020
|
|
6L95
 
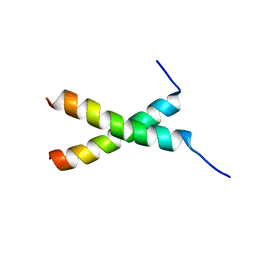 | | transmembrane-domain of Bax | | Descriptor: | Apoptosis regulator BAX | | Authors: | Bo, O.Y, Fujiao, L. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | transmembrane-domain of Bax
To Be Published
|
|
6L99
 
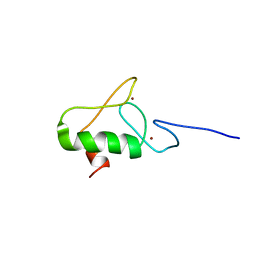 | | Zinc finger of RING finger protein 144A | | Descriptor: | E3 ubiquitin-protein ligase RNF144A, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2019-11-08 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc finger of RING finger protein 144A
To Be Published
|
|
6LAG
 
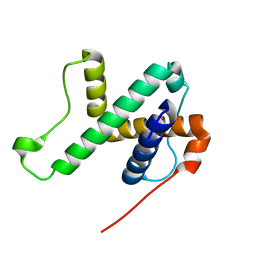 | | Solution structure of SPA-2 SHD | | Descriptor: | Spa2-like protein | | Authors: | Fan, J.S, Wong, J.Y, Zheng, P, Yang, D, Jedd, G. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Spitzenkorper assembly mechanisms reveal conserved features of fungal and metazoan polarity scaffolds.
Nat Commun, 11, 2020
|
|
6LCI
 
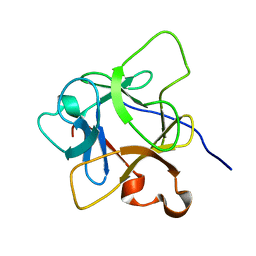 | | Solution structure of mdaA-1 domain | | Descriptor: | mdaA-1 | | Authors: | Nguyen, T.A, Le, S, Lee, M, Fan, J.S, Yang, D, Yan, J, Jedd, G. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Fungal Wound Healing through Instantaneous Protoplasmic Gelation.
Curr.Biol., 31, 2021
|
|
6LEK
 
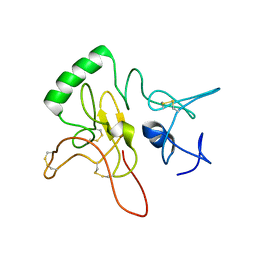 | | Tertiary structure of Barnacle cement protein MrCP20 | | Descriptor: | Cement protein-20k | | Authors: | Mohanram, H. | | Deposit date: | 2019-11-25 | | Release date: | 2020-01-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of Megabalanus rosa Cement Protein 20 revealed by multi-dimensional NMR and molecular dynamics simulations.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 374, 2019
|
|
6LF5
 
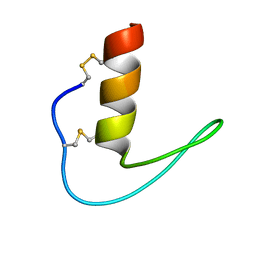 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
6LKF
 
 | |
6LLQ
 
 | | Solution NMR structure of de novo Rossmann2x2 fold with most of the core mutated to valine, R2x2_VAL88 | | Descriptor: | VAL88 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Koga, R, Yamamoto, M, Kosugi, T, Koga, N. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LMR
 
 | | Solution structure of cold shock domain and ssDNA complex | | Descriptor: | DNA (5'-D(P*AP*AP*CP*AP*CP*CP*T)-3'), Y-box-binding protein 1 | | Authors: | Fan, J, Yang, D. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA binding to human YB-1 cold shock domain regulated by phosphorylation.
Nucleic Acids Res., 48, 2020
|
|
6LMS
 
 | |
6LNZ
 
 | |
6LQZ
 
 | | Solution structure of Taf14ET-Sth1EBMC | | Descriptor: | Nuclear protein STH1/NPS1, Transcription initiation factor TFIID subunit 14 | | Authors: | Wu, B, Chen, G, Chen, Y. | | Deposit date: | 2020-01-15 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Taf14 recognizes a common motif in transcriptional machineries and facilitates their clustering by phase separation.
Nat Commun, 11, 2020
|
|
