2MXG
 
 | |
2MXH
 
 | |
2MXJ
 
 | |
2MXK
 
 | |
2MXL
 
 | |
2MXM
 
 | |
2MXN
 
 | | NMR Structure of the mature form of Trypanosoma brucei 1CGrx1 | | Descriptor: | Mono-cysteine glutaredoxin | | Authors: | Sturlese, M, Bertarello, A, Manta, B, Lelli, M, Mammi, S, Comini, M, Bellanda, M. | | Deposit date: | 2015-01-08 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The lineage-specific, intrinsically disordered N-terminal extension of monothiol glutaredoxin 1 from trypanosomes contains a regulatory region.
Sci Rep, 8, 2018
|
|
2MXO
 
 | |
2MXP
 
 | |
2MXQ
 
 | | The solution structure of DEFA1, a highly potent antimicrobial peptide from the horse | | Descriptor: | Paneth cell-specific alpha-defensin 1 | | Authors: | Jung, S, Michalek, M, Shomali, M, Soennichsen, F.D. | | Deposit date: | 2015-01-12 | | Release date: | 2015-04-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure and functional studies of the highly potent equine antimicrobial peptide DEFA1.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
2MXS
 
 | | Solution NMR-structure of the neomycin sensing riboswitch RNA bound to paromomycin | | Descriptor: | PAROMOMYCIN, RNA (27-MER) | | Authors: | Schmidtke, S, Duchardt-Ferner, E, Ohlenschlaeger, O, Gottstein, D, Wohnert, J. | | Deposit date: | 2015-01-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | What a Difference an OH Makes: Conformational Dynamics as the Basis for the Ligand Specificity of the Neomycin-Sensing Riboswitch.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2MXT
 
 | |
2MXU
 
 | | 42-Residue Beta Amyloid Fibril | | Descriptor: | Amyloid beta A4 protein | | Authors: | Xiao, Y, Ma, B, McElheny, D, Parthasarathy, S, Long, F, Hoshi, M, Nussinov, R, Ishii, Y. | | Deposit date: | 2015-01-14 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | A beta (1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
2MXV
 
 | |
2MXW
 
 | |
2MXX
 
 | |
2MXY
 
 | | Solution structure of hnRNP C RRM in complex with 5'-AUUUUUC-3' RNA | | Descriptor: | 5'-R(*AP*UP*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-01-19 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
2MXZ
 
 | |
2MY1
 
 | | Solution structure of Bud31p | | Descriptor: | Pre-mRNA-splicing factor BUD31, ZINC ION | | Authors: | van Roon, A.M, Yang, J, Mathieu, D, Bermel, W, Nagai, K, Neuhaus, D. | | Deposit date: | 2015-01-19 | | Release date: | 2015-03-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | (113) Cd NMR Experiments Reveal an Unusual Metal Cluster in the Solution Structure of the Yeast Splicing Protein Bud31p.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2MY2
 
 | |
2MY3
 
 | |
2MY5
 
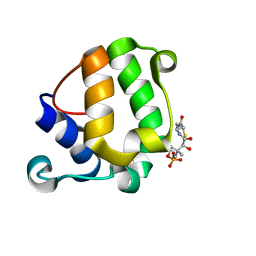 | | Solution Structure of KstB-PCP in kosinostatin biosynthesis | | Descriptor: | N~3~-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-N-(2-sulfanylethyl)-beta-alaninamide, Peptidyl carrier protein | | Authors: | Zhao, B, Lan, W, Wang, C, Tang, G, Cao, C. | | Deposit date: | 2015-01-20 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | 1H and 15N Assigned Chemical Shifts for KstB-PCP
To be Published
|
|
2MY6
 
 | |
2MY7
 
 | |
2MY8
 
 | |
