7K3G
 
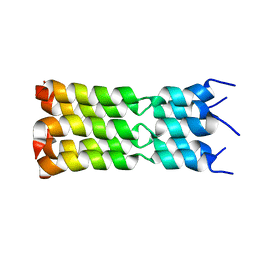 | | SARS-CoV-2 Envelope Protein Transmembrane Domain: Pentameric Structure Determined by Solid-State NMR | | Descriptor: | Envelope small membrane protein | | Authors: | Mandala, V.S, Hong, M, McKay, M.J, Shcherbakov, A.S, Dregni, A.J. | | Deposit date: | 2020-09-11 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7K3S
 
 | |
7K4L
 
 | |
7K6B
 
 | |
7K7A
 
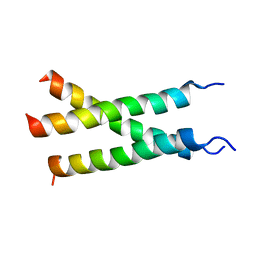 | | Transmembrane structure of TNFR1 | | Descriptor: | Tumor necrosis factor receptor superfamily member 1A | | Authors: | Zhao, L, Chou, J. | | Deposit date: | 2020-09-22 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Diversity and Similarity of Transmembrane Trimerization of TNF Receptors.
Front Cell Dev Biol, 8, 2020
|
|
7K7F
 
 | |
7K7X
 
 | |
7KAA
 
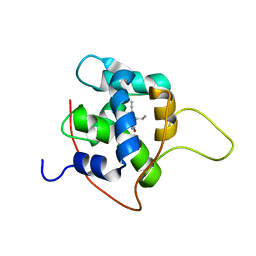 | | NMR solution structures of tirasemtiv drug bound to a fast skeletal troponin C-troponin I complex | | Descriptor: | 6-ethynyl-1-(pentan-3-yl)-1H-imidazo[4,5-b]pyrazin-2-ol, CALCIUM ION, Troponin C, ... | | Authors: | Mercier, P, Li, M.X, Hartman, J.J, Sykes, B.D. | | Deposit date: | 2020-09-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Tirasemtiv Activation of Fast Skeletal Muscle.
J.Med.Chem., 64, 2021
|
|
7KBQ
 
 | |
7KBV
 
 | |
7KBW
 
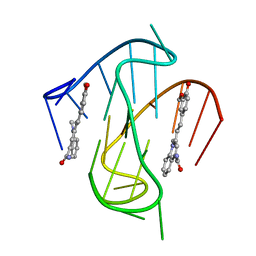 | |
7KBX
 
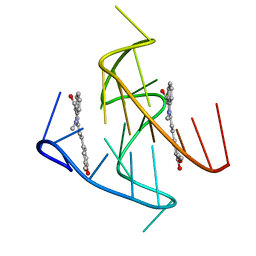 | |
7KDQ
 
 | |
7KIX
 
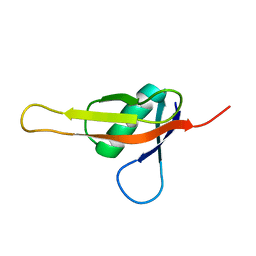 | |
7KLO
 
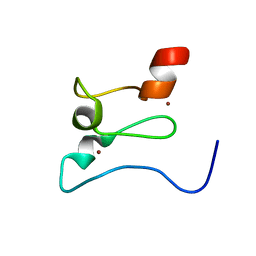 | |
7KLR
 
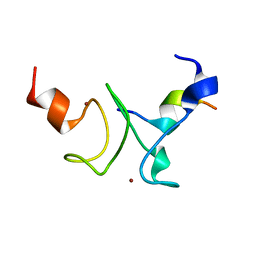 | |
7KN0
 
 | | Structure of the integrin aIIb(W968V)b3 transmembrane complex | | Descriptor: | Integrin alpha-IIb, Integrin beta-3 | | Authors: | Situ, A.J, Kim, J, An, W, Kim, C, Ulmer, T.S. | | Deposit date: | 2020-11-03 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Insight Into Pathological Integrin alpha IIb beta 3 Activation From Safeguarding The Inactive State.
J.Mol.Biol., 433, 2021
|
|
7KNN
 
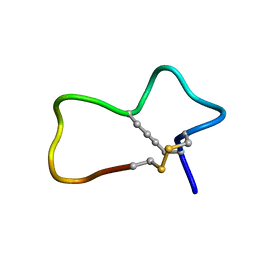 | |
7KNV
 
 | | Solution NMR structure of CDHR3 extracellular domain EC1 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3 | | Authors: | Lee, W, Tonelli, M, Frederick, R.O, Watters, K.E, Markley, J.L, Palmenberg, A.C. | | Deposit date: | 2020-11-06 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Determination of the CDHR3 Rhinovirus-C Binding Domain, EC1
Viruses, 13, 2021
|
|
7KPD
 
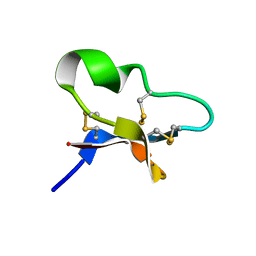 | |
7KRB
 
 | |
7KS6
 
 | | STRUCTURE OF TETRASACCHARIDE BUILDING BLOCK OF A SULFATED FUCAN FROM LYTECHINUS VARIEGATUS | | Descriptor: | 4-O-sulfo-alpha-L-fucopyranose-(1-3)-2,4-di-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose-(1-3)-2-O-sulfo-alpha-L-fucopyranose | | Authors: | Kim, S.B, Thara, R, Aderibigbe, A.O, Doerksen, R.J, Pomin, V.H. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformational properties of l-fucose and the tetrasaccharide building block of the sulfated l-fucan from Lytechinus variegatus.
J.Struct.Biol., 209, 2020
|
|
7KUB
 
 | |
7KUC
 
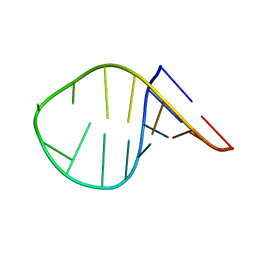 | |
7KUD
 
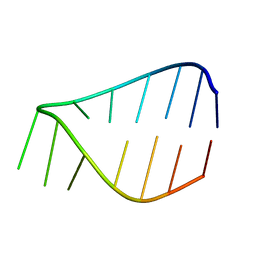 | |
