6GAT
 
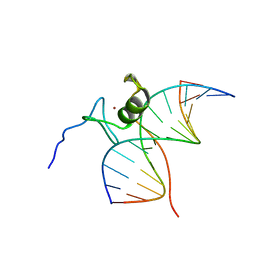 | | SOLUTION NMR STRUCTURE OF THE L22V MUTANT DNA BINDING DOMAIN OF AREA COMPLEXED TO A 13 BP DNA CONTAINING A TGATA SITE, REGULARIZED MEAN STRUCTURE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*GP*AP*TP*AP*GP*AP*GP*AP*C)-3'), DNA (5'-D(*GP*TP*CP*TP*CP*TP*AP*TP*CP*AP*CP*TP*G)-3'), NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Clore, G.M, Starich, M, Wikstrom, M, Gronenborn, A.M. | | Deposit date: | 1997-11-07 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Leu22-->Val mutant AREA DNA binding domain complexed with a TGATAG core element defines a role for hydrophobic packing in the determination of specificity.
J.Mol.Biol., 277, 1998
|
|
6GBD
 
 | |
6GBE
 
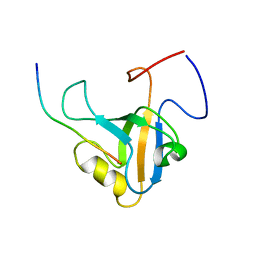 | |
6GBM
 
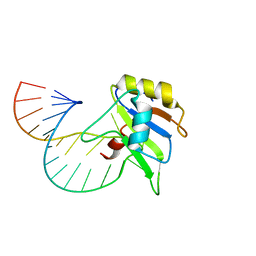 | | Solution structure of FUS-RRM bound to stem-loop RNA | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*GP*AP*UP*UP*AP*CP*AP*AP*UP*UP*CP*UP*AP*UP*UP*UP*GP*CP*C)-3'), RNA-binding protein FUS | | Authors: | Loughlin, F.E, Allain, F.H.-T. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of FUS Bound to RNA Reveals a Bipartite Mode of RNA Recognition with Both Sequence and Shape Specificity.
Mol. Cell, 73, 2019
|
|
6GC3
 
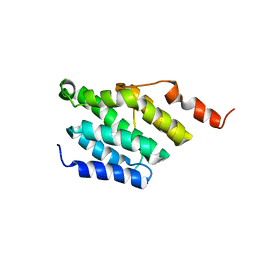 | |
6GCJ
 
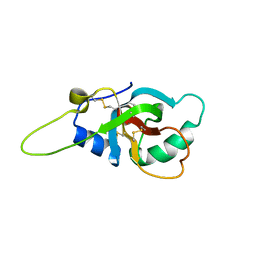 | | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | | Descriptor: | Hydrophobin | | Authors: | Pille, A, Kwan, A, Aimanianda, V, Latge, J.-P, Sunde, M, Guijarro, J.I. | | Deposit date: | 2018-04-18 | | Release date: | 2019-03-27 | | Last modified: | 2019-09-04 | | Method: | SOLUTION NMR | | Cite: | Assembly and disassembly of Aspergillus fumigatus conidial rodlets
Cell Surf, 2019
|
|
6GD5
 
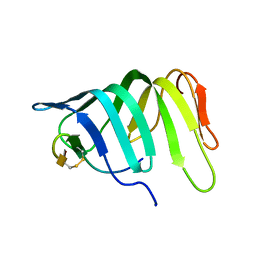 | | The solution structure of the LptA-Thanatin complex | | Descriptor: | Lipopolysaccharide export system protein LptA, Thanatin | | Authors: | Moehle, K, Zerbe, O. | | Deposit date: | 2018-04-22 | | Release date: | 2018-11-28 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport inEscherichia coli.
Sci Adv, 4, 2018
|
|
6GDK
 
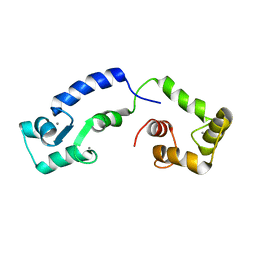 | | Calcium bound form of human calmodulin mutant F141L | | Descriptor: | CALCIUM ION, Calmodulin-1 | | Authors: | Grachov, O, Holt, C, Overgaard, M.T, Wimmer, R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Arrhythmia mutations in calmodulin cause conformational changes that affect interactions with the cardiac voltage-gated calcium channel.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6GDL
 
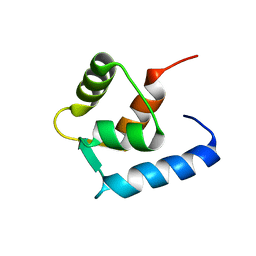 | |
6GDZ
 
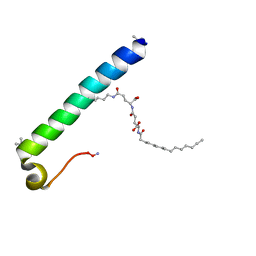 | | exendin-4 based dual GLP-1/glucagon receptor agonist | | Descriptor: | (2~{S})-2-[[(4~{S})-4-(hexadecanoylamino)-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]pentanedioic acid, Exendin-4 | | Authors: | Evers, A, Kurz, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6GE1
 
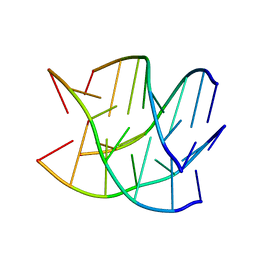 | |
6GE2
 
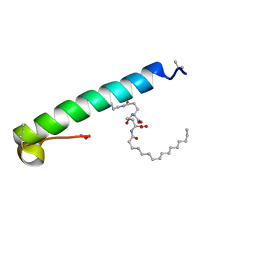 | | exendin-4 based dual GLP-1/glucagon receptor agonist | | Descriptor: | (2~{S})-2-[[(4~{S})-4-(hexadecanoylamino)-5-oxidanyl-5-oxidanylidene-pentanoyl]amino]pentanedioic acid, Exendin-4 | | Authors: | Evers, A, Kurz, M. | | Deposit date: | 2018-04-25 | | Release date: | 2018-06-20 | | Last modified: | 2019-05-08 | | Method: | SOLUTION NMR | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6GF2
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-08 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
6GFT
 
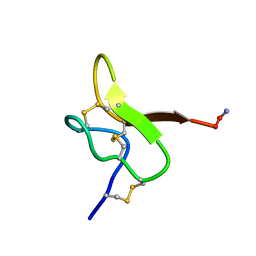 | |
6GGZ
 
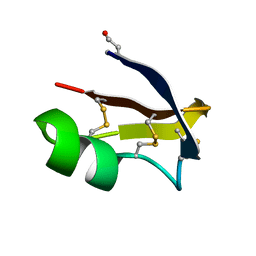 | | NMR structure of the scorpion toxin AmmTx3 | | Descriptor: | Potassium channel toxin alpha-KTx 15.3 | | Authors: | Landon, C, Meudal, H. | | Deposit date: | 2018-05-04 | | Release date: | 2019-01-30 | | Last modified: | 2020-03-11 | | Method: | SOLUTION NMR | | Cite: | Synthesis by native chemical ligation and characterization of the scorpion toxin AmmTx3.
Bioorg. Med. Chem., 27, 2019
|
|
6GH0
 
 | |
6GIF
 
 | |
6GIG
 
 | |
6GIJ
 
 | | NMR structure of temporin B KKG6A in SDS micelles | | Descriptor: | temporinB_KKG6A | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GIK
 
 | | NMR structure of temporin B L1FK in SDS micelles | | Descriptor: | temporinB_L1FK | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GIL
 
 | | NMR structure of temporin B in SDS micelles | | Descriptor: | Temporin-B | | Authors: | Manzo, G, Mason, J.A. | | Deposit date: | 2018-05-12 | | Release date: | 2018-06-13 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Minor sequence modifications in temporin B cause drastic changes in antibacterial potency and selectivity by fundamentally altering membrane activity.
Sci Rep, 9, 2019
|
|
6GMS
 
 | | Solution NMR structure of the major type IV pilin PpdD from enterohemorrhagic Escherichia coli (EHEC) | | Descriptor: | Prepilin peptidase-dependent protein D | | Authors: | Amorim, G.C, Bardiaux, B, Luna-Rico, A, Zeng, W, Guilvout, I, Egelman, E, Nilges, M, Francetic, O, Izadi-Pruneyre, N. | | Deposit date: | 2018-05-28 | | Release date: | 2019-05-15 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure and Assembly of the Enterohemorrhagic Escherichia coli Type 4 Pilus.
Structure, 27, 2019
|
|
6GMY
 
 | | Tc-DNA/RNA duplex | | Descriptor: | RNA (5'-R(*GP*UP*AP*AP*GP*CP*CP*GP*AP*G)-3'), Tc-DNA (5'-(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GN4
 
 | | tc-DNA/tc-DNA duplex | | Descriptor: | Tc-DNA (5'-D(*(TCJ)P*(TTK)P*(TCJ)P*(TCS)P*(TCS)P*(TCJ)P*(TTK)P*(TTK)P*(TCY)P*(TCJ))-3'), Tc-DNA (5'-D(*(TCS)P*(TTK)P*(TCY)P*(TCY)P*(TCS)P*(TCJ)P*(TCJ)P*(TCS)P*(TCY)P*(TCS))-3') | | Authors: | Istrate, A, Johannsen, S, Istrate, A, Sigel, R.K.O, Leumann, C. | | Deposit date: | 2018-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of tricyclo-DNA containing duplexes: insight into enhanced thermal stability and nuclease resistance.
Nucleic Acids Res., 47, 2019
|
|
6GNZ
 
 | |
