5MML
 
 | | HYL-20k | | Descriptor: | GLY-ILE-LEU-SER-SER-LEU-TRP-LYS-LYS-LEU-LYS-LYS-ILE-ILE-ALA-LYS | | Authors: | Hexnerova, R. | | Deposit date: | 2016-12-10 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | How proteases from Enterococcus faecalis contribute to its resistance to short alpha-helical antimicrobial peptides.
Pathog Dis, 75, 2017
|
|
5MMU
 
 | | NMR solution structure of the major apple allergen Mal d 1 | | Descriptor: | Major allergen Mal d 1 | | Authors: | Ahammer, L, Grutsch, S, Kamenik, A.S, Liedl, K.R, Tollinger, M. | | Deposit date: | 2016-12-12 | | Release date: | 2017-02-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Major Apple Allergen Mal d 1.
J. Agric. Food Chem., 65, 2017
|
|
5MN3
 
 | |
5MNW
 
 | |
5MOU
 
 | | NMR spatial structure of scorpion alpha-like toxin BeM9 | | Descriptor: | Alpha-mammal toxin BeM9 | | Authors: | Mineev, K.S, Kuldushev, N.A, Berkut, A.A, Grishin, E.V, Vassilevski, A.A, Arseniev, A.S. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Refined structure of BeM9 reveals arginine hand, an overlooked structural motif in scorpion toxins affecting sodium channels.
Proteins, 86, 2018
|
|
5MPG
 
 | |
5MPL
 
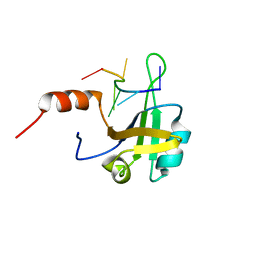 | | hnRNP A1 RRM2 in complex with 5'-UCAGUU-3' RNA | | Descriptor: | Heterogeneous nuclear ribonucleoprotein A1, RNA UCAGUU | | Authors: | Barraud, P, Allain, F.H.-T. | | Deposit date: | 2016-12-16 | | Release date: | 2017-07-05 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Tandem hnRNP A1 RNA recognition motifs act in concert to repress the splicing of survival motor neuron exon 7.
Elife, 6, 2017
|
|
5MQX
 
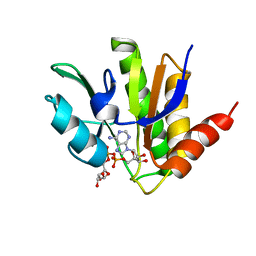 | | NMR solution structure of macro domain from Venezuelan equine encephalitis virus(VEEV) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein3 | | Authors: | Makrynitsa, G.I, Ntonti, D, Marousis, K.D, Matsoukas, M.T, Papageorgiou, N, Coutard, B, Bentrop, D, Spyroulias, G.A. | | Deposit date: | 2016-12-21 | | Release date: | 2018-07-04 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Conformational plasticity of the VEEV macro domain is important for binding of ADP-ribose.
J.Struct.Biol., 206, 2019
|
|
5MRG
 
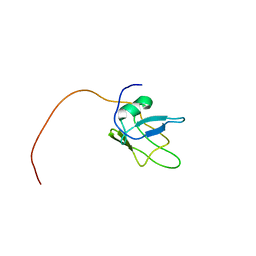 | | Solution structure of TDP-43 (residues 1-102) | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F.E, Laurents, D.V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.
J. Biol. Chem., 292, 2017
|
|
5MS9
 
 | |
5MSL
 
 | |
5MTA
 
 | |
5MTG
 
 | |
5MTI
 
 | | Bamb_5917 Acyl-Carrier Protein | | Descriptor: | Phosphopantetheine-binding protein | | Authors: | Gallo, A, Kosol, S, Griffiths, D, Masschelein, J, Alkhalaf, L, Smith, H, Valentic, T, Tsai, S, Challis, G, Lewandowski, J.R. | | Deposit date: | 2017-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for chain release from the enacyloxin polyketide synthase
Nat.Chem., 2019
|
|
5MVB
 
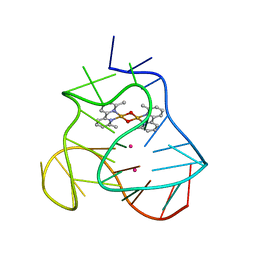 | | Solution structure of a human G-Quadruplex hybrid-2 form in complex with a Gold-ligand. | | Descriptor: | DNA (26-MER), POTASSIUM ION, bis(m2-Oxo)-bis(2-methyl-2,2'-bipyridine)-di-gold(iii) | | Authors: | Wirmer-Bartoschek, J, Jonker, H.R.A, Bendel, L.E, Gruen, T, Bazzicalupi, C, Messori, L, Gratteri, P, Schwalbe, H. | | Deposit date: | 2017-01-16 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of a Ligand/Hybrid-2-G-Quadruplex Complex Reveals Rearrangements that Affect Ligand Binding.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MWQ
 
 | |
5MWV
 
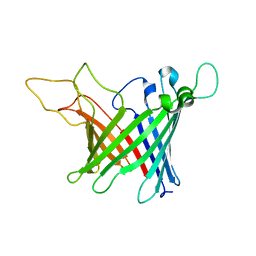 | | Solid-state NMR Structure of outer membrane protein G in lipid bilayers | | Descriptor: | Outer membrane protein G | | Authors: | Retel, J.S, Nieuwkoop, A.J, Hiller, M, Higman, V.A, Barbet-Massin, E, Stanek, J, Andreas, L.B, Franks, W.T, van Rossum, B.-J, Vinothkumar, K.R, Handel, L, de Palma, G.G, Bardiaux, B, Pintacuda, G, Emsley, L, Kuelbrandt, W, Oschkinat, H. | | Deposit date: | 2017-01-20 | | Release date: | 2017-12-27 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Structure of outer membrane protein G in lipid bilayers.
Nat Commun, 8, 2017
|
|
5MWW
 
 | | Sigma1.1 domain of sigmaA from Bacillus subtilis | | Descriptor: | RNA polymerase sigma factor SigA | | Authors: | Zachrdla, M, Padrta, P, Rabatinova, A, Sanderova, H, Barvik, I, Krasny, L, Zidek, L. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution structure of domain 1.1 of the sigma (A) factor from Bacillus subtilis is preformed for binding to the RNA polymerase core.
J. Biol. Chem., 292, 2017
|
|
5MXL
 
 | |
5MXS
 
 | |
5MXT
 
 | |
5MYE
 
 | |
5N14
 
 | |
5N2O
 
 | | Structure Of P63 SAM Domain L514F Mutant Causative Of AEC Syndrome | | Descriptor: | Tumor protein 63 | | Authors: | Rinnenthal, J, Wuerz, J.M, Osterburg, C, Guentert, P, Doetsch, V. | | Deposit date: | 2017-02-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Protein aggregation of the p63 transcription factor underlies severe skin fragility in AEC syndrome.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5N5A
 
 | |
