2K5U
 
 | |
2K5V
 
 | | SOLUTION NMR STRUCTURE OF the second OB-fold domain of replication protein A from Methanococcus maripaludis. NORTHEAST STRUCTURAL GENOMICS TARGET MrR110B. | | Descriptor: | Replication protein A | | Authors: | Aramini, J.M, Maglaqui, M, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.VT, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION NMR STRUCTURE OF the second OB-fold domain of replication protein A from Methanococcus maripaludis. NORTHEAST STRUCTURAL GENOMICS TARGET MrR110B.
To be Published
|
|
2K5W
 
 | | Solution NMR Structure of Putative Lipoprotein from Bacillus cereus Ordered Locus BC_2438. Northeast Structural Genomics Target BcR103A. | | Descriptor: | Hypothetical Cytosolic Protein BcR103A | | Authors: | Conover, K.M, Swapna, G, Rossi, P, Wang, D, Janjua, H, Owens, L, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Putative Lipoprotein from Bacillus cereus Ordered Locus BC_2438. Northeast Structural Genomics Target BcR103A.
To be Published
|
|
2K5X
 
 | |
2K5Z
 
 | |
2K60
 
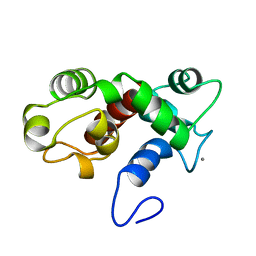 | | NMR structure of calcium-loaded STIM1 EF-SAM | | Descriptor: | CALCIUM ION, PROTEIN (Stromal interaction molecule 1) | | Authors: | Stathopulos, P.B, Ikura, M. | | Deposit date: | 2008-07-02 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into STIM1-mediated initiation of store-operated calcium entry.
Cell(Cambridge,Mass.), 135, 2008
|
|
2K61
 
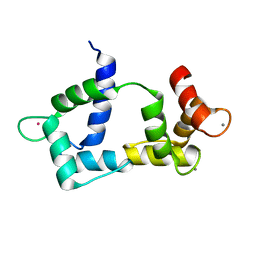 | | Solution structure of CaM complexed to DAPk peptide | | Descriptor: | CALCIUM ION, Calmodulin, TERBIUM(III) ION | | Authors: | Bertini, I, Luchinat, C, Parigi, G, Yuan, J. | | Deposit date: | 2008-07-02 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Accurate solution structures of proteins from X-ray data and a minimal set of NMR data: calmodulin-peptide complexes as examples.
J.Am.Chem.Soc., 131, 2009
|
|
2K62
 
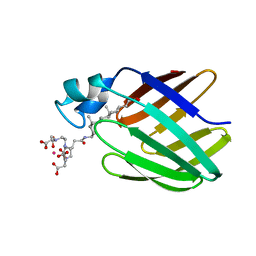 | | NMR solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based Gd(III)-chelate | | Descriptor: | (3alpha,5alpha,8alpha)-3-[(N,N-bis{2-[bis(carboxymethyl)amino]ethyl}-L-gamma-glutamyl)amino]cholan-24-oic acid, Liver fatty acid-binding protein, YTTERBIUM (III) ION | | Authors: | Tomaselli, S, Zanzoni, S, Ragona, L, Gianolio, E, Aime, S, Assfalg, M, Molinari, H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the supramolecular adduct between a liver cytosolic bile acid binding protein and a bile acid-based gadolinium(III)-chelate, a potential hepatospecific magnetic resonance imaging contrast agent.
J.Med.Chem., 51, 2008
|
|
2K65
 
 | |
2K66
 
 | |
2K67
 
 | |
2K68
 
 | |
2K69
 
 | |
2K6A
 
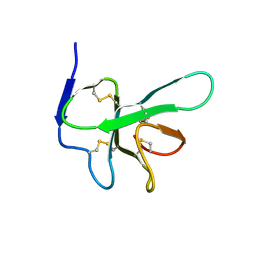 | | Solution structure of EAS D15 truncation mutant | | Descriptor: | Hydrophobin | | Authors: | Kwan, A.H. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Cys3-Cys4 loop of the hydrophobin EAS is not required for rodlet formation and surface activity.
J.Mol.Biol., 382, 2008
|
|
2K6B
 
 | |
2K6D
 
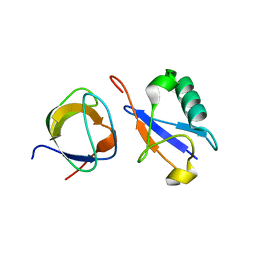 | | CIN85 Sh3-C domain in complex with ubiquitin | | Descriptor: | SH3 domain-containing kinase-binding protein 1, Ubiquitin | | Authors: | Forman-Kay, J, Bezsonova, I. | | Deposit date: | 2008-07-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Interactions between the three CIN85 SH3 domains and ubiquitin: implications for CIN85 ubiquitination.
Biochemistry, 47, 2008
|
|
2K6G
 
 | |
2K6H
 
 | |
2K6I
 
 | | The domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H1-47 | | Descriptor: | Uncharacterized protein MJ0223 | | Authors: | Biukovic, N, Gayen, S, Pervushin, K, Gruber, G, Biukovic, G. | | Deposit date: | 2008-07-09 | | Release date: | 2009-07-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Domain features of the peripheral stalk subunit H of the methanogenic A1AO ATP synthase and the NMR solution structure of H(1-47).
Biophys.J., 97, 2009
|
|
2K6L
 
 | | The solution structure of XACb0070 from Xanthonomas axonopodis pv citri reveals this new protein is a member of the RHH family of transcriptional repressors | | Descriptor: | Putative uncharacterized protein | | Authors: | Gallo, M, Cicero, D.O, Amata, I, Eliseo, T, Paci, M, Spisni, A, Ferrari, E, Pertinhez, T.A, Farah, C.S. | | Deposit date: | 2008-07-11 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure reveals that XACb0070 from the plant pathogen Xanthomonas citri belongs to the RHH superfamily of bacterial DNA-binding proteins
To be Published
|
|
2K6M
 
 | |
2K6N
 
 | |
2K6O
 
 | | Human LL-37 Structure | | Descriptor: | Cathelicidin antimicrobial peptide | | Authors: | Wang, G. | | Deposit date: | 2008-07-14 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles
J.Biol.Chem., 283, 2008
|
|
2K6P
 
 | | Solution Structure of hypothetical protein, HP1423 | | Descriptor: | Uncharacterized protein HP_1423 | | Authors: | Kim, J, Park, S, Lee, K, Son, W, Sohn, N, Lee, B. | | Deposit date: | 2008-07-15 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of hypothetical protein HP1423 (Y1423_HELPY) reveals the presence of alphaL motif related to RNA binding
Proteins, 75, 2009
|
|
2K6Q
 
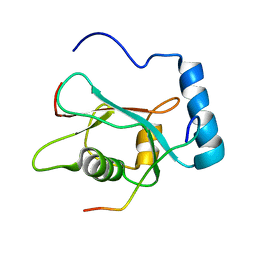 | | LC3 p62 complex structure | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B, p62_peptide from Sequestosome-1 | | Authors: | Noda, N, Kumeta, H, Nakatogawa, H, Satoo, K, Adachi, W, Ishii, J, Fujioka, Y, Ohsumi, Y, Inagaki, F. | | Deposit date: | 2008-07-17 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of target recognition by ATG8/LC3 during selective autophagy
To be Published
|
|
