2JZI
 
 | |
2JZJ
 
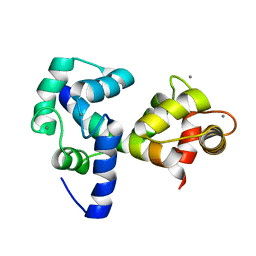
 | | Structure of CrCVNH (C. richardii CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZK
 
 | | Structure of TbCVNH (T. BORCHII CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZL
 
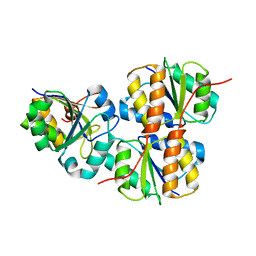
 | | Structure of NcCVNH (N. CRASSA CVNH) | | Descriptor: | Cyanovirin-N homolog | | Authors: | Koharudin, L.M.I, Viscomi, A.R, Jee, J, Ottonello, S, Gronenborn, A.M. | | Deposit date: | 2008-01-09 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The evolutionarily conserved family of cyanovirin-N homologs: structures and carbohydrate specificity.
Structure, 16, 2008
|
|
2JZM
 
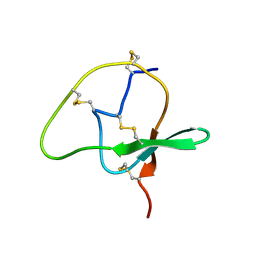 | |
2JZN
 
 | |
2JZO
 
 | |
2JZQ
 
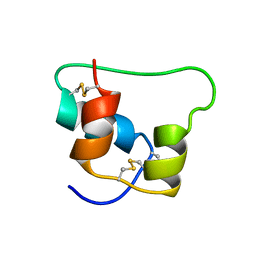 | | Design of an Active Ultra-Stable Single-Chain Insulin Analog 20 Structures | | Descriptor: | Insulin | | Authors: | Hua, Q.X, Nakarawa, S, Jia, W.H, Huang, K, Philips, N.F, Hu, S.Q, Weiss, M.A. | | Deposit date: | 2008-01-11 | | Release date: | 2008-02-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications.
J.Biol.Chem., 283, 2008
|
|
2JZR
 
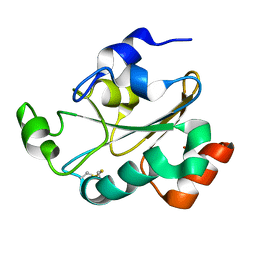 | | Solution structure of the oxidized form (Cys67-Cys70) of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2JZS
 
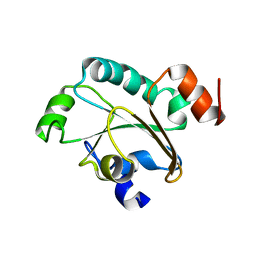 | | Solution structure of the reduced form of the N-terminal domain of PilB from N. meningitidis. | | Descriptor: | Peptide methionine sulfoxide reductase msrA/msrB | | Authors: | Quinternet, M, Tsan, P, Neiers, F, Beaufils, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-01-15 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of the reduced and oxidized forms of the N-terminal domain of PilB from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
2JZT
 
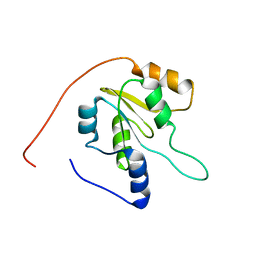 | | Solution NMR structure of Q8ZP25_SALTY from Salmonella typhimurium. Northeast Structural Genomics Consortium target StR70 | | Descriptor: | Putative thiol-disulfide isomerase and thioredoxin | | Authors: | Parish, D, Liu, G, Shen, Y, Ho, C, Cunningham, K, Xiao, R, Swapna, G.V.T, Acton, T, Bansal, S, Prestegard, J.H, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-16 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Protein chaperones Q8ZP25_SALTY from Salmonella typhimurium and HYAE_ECOLI from Escherichia coli exhibit thioredoxin-like structures despite lack of canonical thioredoxin active site sequence motif.
J.STRUCT.FUNCT.GENOM., 9, 2008
|
|
2JZV
 
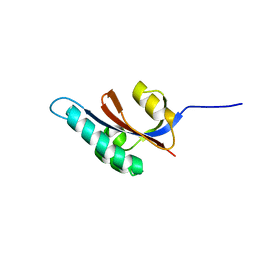 | | Solution structure of S. aureus PrsA-PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Seppala, R, Tossavainen, H, Heikkinen, S, Koskela, H, Kontinen, V, Permi, P. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA - Implications for the catalytic mechanism of parvulins.
Bmc Struct.Biol., 9, 2009
|
|
2JZW
 
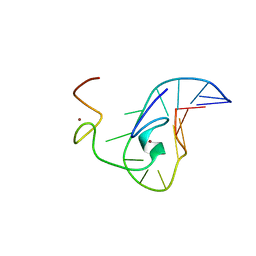 | | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription | | Descriptor: | DNA (5'-D(*DGP*DTP*DCP*DCP*DCP*DTP*DGP*DTP*DTP*DCP*DGP*DGP*DGP*DC)-3'), HIV-1 nucleocapsid protein NCp7(12-55), ZINC ION | | Authors: | Bourbigot, S, Ramalanjaona, N, Salgado, G.F.J, Mely, Y, Roques, B.P, Bouaziz, S, Morellet, N. | | Deposit date: | 2008-01-21 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | How the HIV-1 nucleocapsid protein binds and destabilises the (-)primer binding site during reverse transcription.
J.Mol.Biol., 383, 2008
|
|
2JZX
 
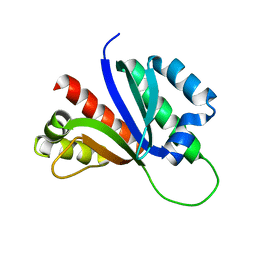 | | PCBP2 KH1-KH2 domains | | Descriptor: | Poly(rC)-binding protein 2 | | Authors: | Du, Z, Fenn, S, Tjhen, R, James, T. | | Deposit date: | 2008-01-21 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the first and second KH domains of human poly-C binding protein-2 reveals insights into its regulatory mechanisms
To be Published
|
|
2JZY
 
 | |
2JZZ
 
 | |
2K00
 
 | |
2K01
 
 | |
2K02
 
 | |
2K03
 
 | |
2K04
 
 | |
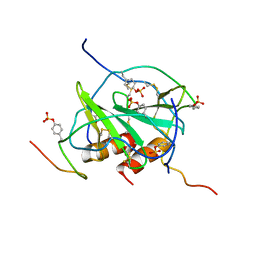
2K05
 
 | |
2K06
 
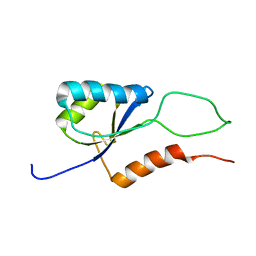 | |
2K07
 
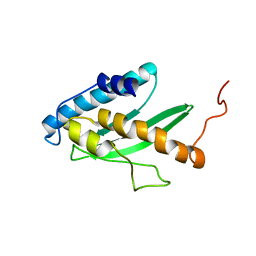 | | Solution NMR structure of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1). Northeast Structural Genomics Consortium target HR41 | | Descriptor: | Ufm1-conjugating enzyme 1 | | Authors: | Liu, G, Eletsky, A, Atreya, H.S, Aramini, J.M, Xiao, R, Acton, T, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-01-25 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR and X-RAY structures of human E2-like ubiquitin-fold modifier conjugating enzyme 1 (UFC1) reveal structural and functional conservation in the metazoan UFM1-UBA5-UFC1 ubiquination pathway.
J.STRUCT.FUNCT.GENOM., 10, 2009
|
|
2K0A
 
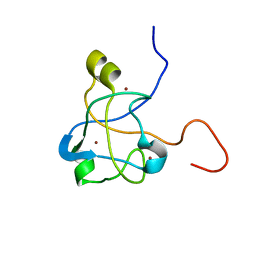 | | 1H, 15N and 13C chemical shift assignments for Rds3 protein | | Descriptor: | Pre-mRNA-splicing factor RDS3, ZINC ION | | Authors: | Loening, N, van Roon, A, Yang, J, Nagai, K, Neuhaus, D. | | Deposit date: | 2008-01-31 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the U2 snRNP protein Rds3p reveals a knotted zinc-finger motif.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
