5Z4E
 
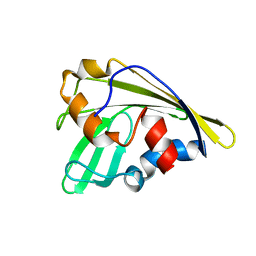 | | An anthrahydroquino-Gama-pyrone synthase Txn09 | | Descriptor: | TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2018-12-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z4F
 
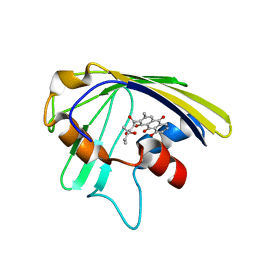 | | An anthrahydroquino-Gama-pyrone synthase Txn09 complexed with SUM | | Descriptor: | 1,8-dihydroxy-2-[(4R)-4-hydroxy-4-methyl-3-oxohexanoyl]-3-methylanthracene-9,10-dione, TxnO9 | | Authors: | Song, Y.J, Cao, C.Y. | | Deposit date: | 2018-01-11 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Enzymology of Anthraquinone-gamma-Pyrone Ring Formation in Complex Aromatic Polyketide Biosynthesis.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Z55
 
 | |
5Z5Q
 
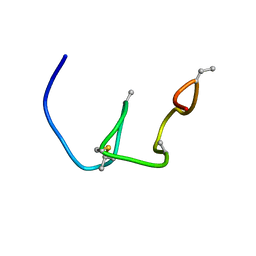 | | Nukacin ISK-1 in active state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
5Z5R
 
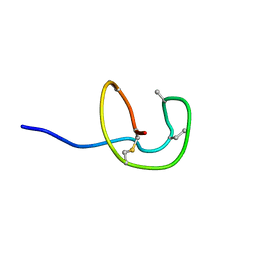 | | Nukacin ISK-1 in inactive state | | Descriptor: | Lantibiotic nukacin | | Authors: | Kohda, D, Fujinami, D. | | Deposit date: | 2018-01-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | The lantibiotic nukacin ISK-1 exists in an equilibrium between active and inactive lipid-II binding states.
Commun Biol, 1, 2018
|
|
5Z5W
 
 | | VFR12 in complex with LPS micelles | | Descriptor: | Peptide from Prothrombin | | Authors: | Saravanan, R, Schmidtchen, A. | | Deposit date: | 2018-01-21 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for endotoxin neutralisation and anti-inflammatory activity of thrombin-derived C-terminal peptides.
Nat Commun, 9, 2018
|
|
5Z5X
 
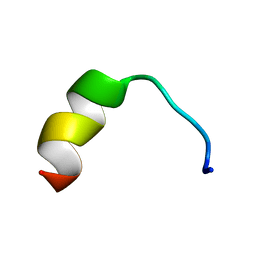 | | HVF18 in complex with LPS micelles | | Descriptor: | Peptide from Prothrombin | | Authors: | Saravanan, R, Schmidtchen, A. | | Deposit date: | 2018-01-21 | | Release date: | 2018-03-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for endotoxin neutralisation and anti-inflammatory activity of thrombin-derived C-terminal peptides.
Nat Commun, 9, 2018
|
|
5Z80
 
 | | Solution structure for the 1:1 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-30 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
5Z8F
 
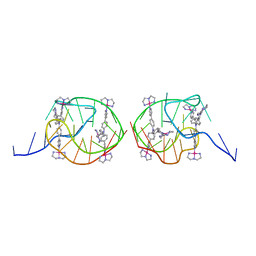 | | Solution structure for the unique dimeric 4:2 complex of a platinum(II)-based tripod bound to a hybrid-1 human telomeric G-quadruplex | | Descriptor: | 4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]-N,N-bis[4-[1-(2,5,8-triazonia-1$l^4-platinabicyclo[3.3.0]octan-1-yl)pyridin-1-ium-4-yl]phenyl]aniline, G-quadruplex DNA (26-MER) | | Authors: | Liu, W.T, Zhong, Y.F, Liu, L.Y, Zeng, W.J, Wang, F.Y, Yang, D.Z, Mao, Z.W. | | Deposit date: | 2018-01-31 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structures of multiple G-quadruplex complexes induced by a platinum(II)-based tripod reveal dynamic binding
Nat Commun, 9, 2018
|
|
5Z8I
 
 | |
5Z8Q
 
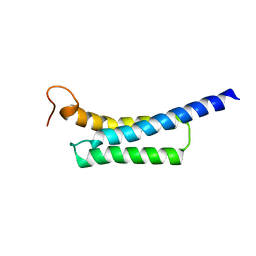 | |
5Z9C
 
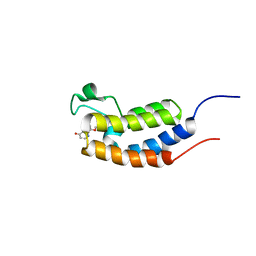 | |
5ZAU
 
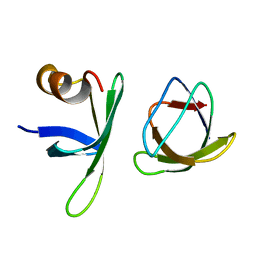 | |
5ZAZ
 
 | |
5ZB6
 
 | |
5ZC6
 
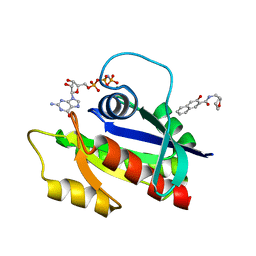 | | Solution structure of H-RasT35S mutant protein in complex with KBFM123 | | Descriptor: | 3-oxidanyl-~{N}-[[(2~{R})-oxolan-2-yl]methyl]naphthalene-2-carboxamide, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Matsumoto, S, Hayashi, Y, Hiraga, T, Matsuo, K, Kataoka, T. | | Deposit date: | 2018-02-15 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular Basis for Allosteric Inhibition of GTP-Bound H-Ras Protein by a Small-Molecule Compound Carrying a Naphthalene Ring
Biochemistry, 57, 2018
|
|
5ZCN
 
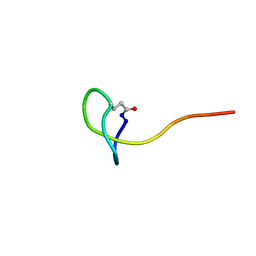 | | Solution NMR structure of a new lasso peptide brevunsin | | Descriptor: | brevunsin | | Authors: | Hemmi, H, Kodani, S, Miyake, Y, Kaweewan, I, Nakagawa, H. | | Deposit date: | 2018-02-19 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Heterologous production of a new lasso peptide brevunsin in Sphingomonas subterranea
J. Ind. Microbiol. Biotechnol., 45, 2018
|
|
5ZCZ
 
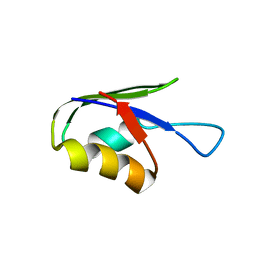 | | Solution structure of the T. Thermophilus HB8 TTHA1718 protein in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | Heavy metal binding protein | | Authors: | Tanaka, T, Teppei, I, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZD0
 
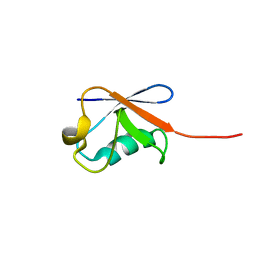 | | Solution structure of human ubiquitin with three alanine mutations in living eukaryotic cells by in-cell NMR spectroscopy | | Descriptor: | ubiquitin | | Authors: | Tanaka, T, Ikeya, T, Kamoshida, H, Mishima, M, Shirakawa, M, Guentert, P, Ito, Y. | | Deposit date: | 2018-02-22 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-Resolution Protein 3D Structure Determination in Living Eukaryotic Cells.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
5ZEV
 
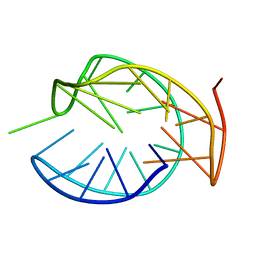 | |
5ZEW
 
 | |
5ZFO
 
 | | NMR structure of IRD12 from Capsicum annum. | | Descriptor: | Pin-II type proteinase inhibitor 38 | | Authors: | Gartia, J, Barnwal, R.P, Chary, K.V.R. | | Deposit date: | 2018-03-06 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | NMR structure and dynamics of inhibitory repeat domain variant 12, a plant protease inhibitor from Capsicum annuum, and its structural relationship to other plant protease inhibitors.
J.Biomol.Struct.Dyn., 2019
|
|
5ZGG
 
 | | NMR structure of p75NTR transmembrane domain in complex with NSC49652 | | Descriptor: | (2E)-1-(2-hydroxyphenyl)-3-(pyridin-3-yl)prop-2-en-1-one, Tumor necrosis factor receptor superfamily member 16 | | Authors: | Lin, Z, Ibanez, C. | | Deposit date: | 2018-03-08 | | Release date: | 2019-03-13 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule Targeting the Transmembrane Domain of Death Receptor p75NTRInduces Melanoma Cell Death and Reduces Tumor Growth.
Cell Chem Biol, 25, 2018
|
|
5ZKV
 
 | | Solution structure of molten globule state of L94G mutant of horse cytochrome-c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Naiyer, A, Islam, A, Hassan, M.I, Sundd, M, Ahmad, F. | | Deposit date: | 2018-03-26 | | Release date: | 2019-05-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of molten globule state of L94G mutant of horse cytochrome-c
To Be Published
|
|
5ZLD
 
 | |
