2MKL
 
 | |
2MKM
 
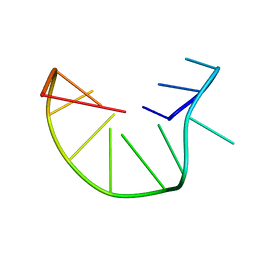 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2MKN
 
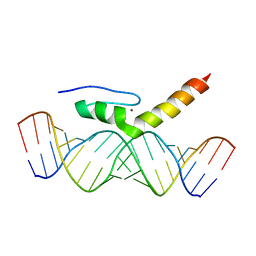 | | Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and dsRNA | | Descriptor: | RNA (5'-R(*GP*CP*CP*GP*UP*GP*GP*UP*CP*UP*GP*GP*UP*GP*GP*CP*CP*GP*G)-3'), RNA (5'-R(P*CP*CP*GP*GP*CP*CP*AP*CP*CP*AP*GP*AP*CP*CP*AP*CP*GP*GP*C)-3'), ZINC ION, ... | | Authors: | Wright, P, Dyson, J, Burge, R, Martinez-Yamout, M. | | Deposit date: | 2014-02-10 | | Release date: | 2014-03-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Characterization of Interactions between the Double-Stranded RNA-Binding Zinc Finger Protein JAZ and Nucleic Acids.
Biochemistry, 53, 2014
|
|
2MKO
 
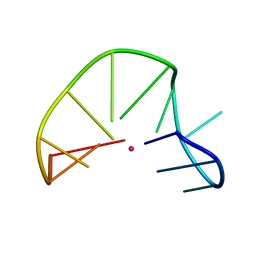 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2MKP
 
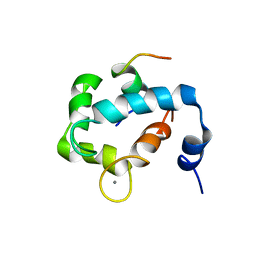 | | N domain of cardiac troponin C bound to the switch fragment of fast skeletal troponin I at pH 6 | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles, ... | | Authors: | Robertson, I.M, Pineda-Sanabria, S.E, Holmes, P.C, Sykes, B.D. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Conformation of the critical pH sensitive region of troponin depends upon a single residue in troponin I.
Arch.Biochem.Biophys., 552-553, 2014
|
|
2MKR
 
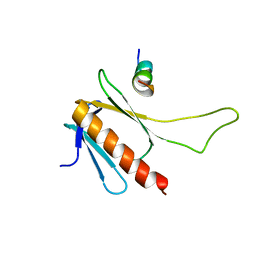 | | Structural Characterization of a Complex Between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 subunit of TFIIH. | | Descriptor: | Epstein-Barr nuclear antigen 2, RNA polymerase II transcription factor B subunit 1 | | Authors: | Chabot, P.R, Raiola, L, Lussier-Price, M, Morse, T, Arseneault, G, Archambault, J, Omichinski, J. | | Deposit date: | 2014-02-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of a Complex between the Acidic Transactivation Domain of EBNA2 and the Tfb1/p62 Subunit of TFIIH.
Plos Pathog., 10, 2014
|
|
2MKS
 
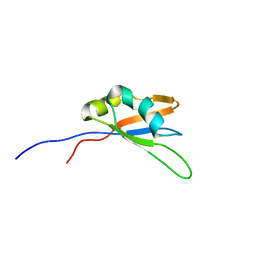 | |
2MKV
 
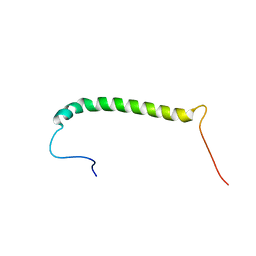 | |
2MKW
 
 | |
2MKX
 
 | |
2MKY
 
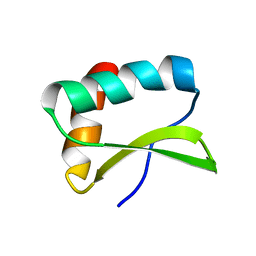 | |
2MKZ
 
 | |
2ML1
 
 | |
2ML2
 
 | |
2ML3
 
 | |
2ML5
 
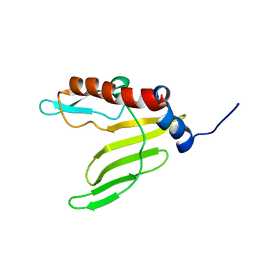 | |
2ML6
 
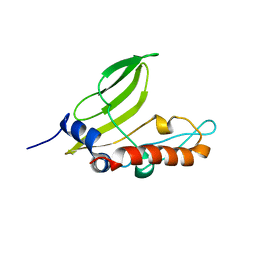 | | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492 | | Descriptor: | Uncharacterized protein | | Authors: | Wallmann, A, Dutta, S.K, Serrano, P, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-02-19 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structure of protein ZP_02069618.1 from Bacteroides uniformis ATCC 8492.
To be Published
|
|
2ML7
 
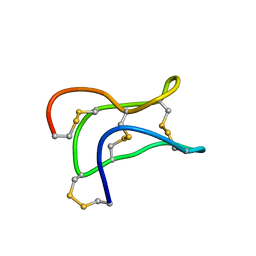 | | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng | | Descriptor: | Specific abundant protein 3 | | Authors: | Wang, S, Nguyen, K, Luo, S, Tam, J, Yang, D. | | Deposit date: | 2014-02-20 | | Release date: | 2015-03-04 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Ginsentides: Characterization, Structure and Application of a New Class of Highly Stable Cystine Knot Peptides in Ginseng
To be Published
|
|
2ML8
 
 | |
2ML9
 
 | |
2MLA
 
 | |
2MLB
 
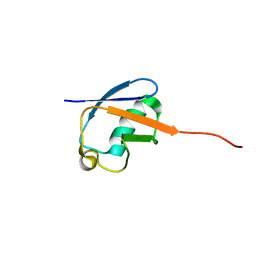 | | NMR solution structure of a computational designed protein based on template of human erythrocytic ubiquitin | | Descriptor: | redesigned ubiquitin | | Authors: | Xiong, P, Wang, M, Zhang, J, Chen, Q, Liu, H. | | Deposit date: | 2014-02-21 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Protein design with a comprehensive statistical energy function and boosted by experimental selection for foldability
Nat Commun, 5, 2014
|
|
2MLD
 
 | |
2MLE
 
 | | NMR structure of the C-domain of troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Robertson, I.M, Baryshnikova, O.K, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
2MLF
 
 | | NMR structure of the dilated cardiomyopathy mutation G159D in troponin C bound to the anchoring region of troponin I | | Descriptor: | CALCIUM ION, Troponin C, slow skeletal and cardiac muscles | | Authors: | Baryshnikova, O.K, Robertson, I.M, Mercier, P, Sykes, B.D. | | Deposit date: | 2014-02-26 | | Release date: | 2014-03-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The dilated cardiomyopathy G159D mutation in cardiac troponin C weakens the anchoring interaction with troponin I.
Biochemistry, 47, 2008
|
|
