7KW8
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis | | Descriptor: | tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KW9
 
 | | NMR Structure of a tRNA 2'-phosphotransferase from Runella slithyformis in complex with NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, tRNA 2'-phosphotransferase | | Authors: | Alphonse, S, Dantuluri, S, Banerjee, A, Shuman, S, Ghose, R. | | Deposit date: | 2020-11-30 | | Release date: | 2021-10-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of Runella slithyformis RNA 2'-phosphotransferase Tpt1 provide insights into NAD+ binding and specificity.
Nucleic Acids Res., 49, 2021
|
|
7KWI
 
 | |
7KWL
 
 | |
7KWR
 
 | |
7KXI
 
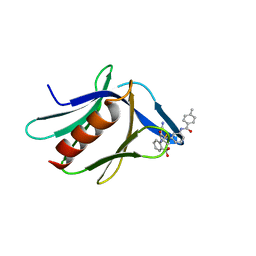 | | Structure of XL5-ligated hRpn13 Pru domain | | Descriptor: | 2-{[(2S)-2-cyano-3-{3-[(4-methylbenzene-1-carbonyl)amino]phenyl}propanoyl]amino}benzoic acid, Proteasomal ubiquitin receptor ADRM1 | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Structure-guided bifunctional molecules hit a DEUBAD-lacking hRpn13 species upregulated in multiple myeloma.
Nat Commun, 12, 2021
|
|
7KYZ
 
 | |
7L2G
 
 | |
7L51
 
 | |
7L53
 
 | |
7L54
 
 | |
7L55
 
 | |
7L6K
 
 | | ApoL1 N-terminal domain | | Descriptor: | Apolipoprotein L1 | | Authors: | Holliday, M.J, Ultsch, M, Moran, P, Fairbrother, W.J, Kirchhofer, D. | | Deposit date: | 2020-12-23 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of the ApoL1 and ApoL2 N-terminal domains reveal a non-classical four-helix bundle motif.
Commun Biol, 4, 2021
|
|
7L7A
 
 | | Solution Structure of NuxVA | | Descriptor: | NuxVA | | Authors: | Mari, F, Dovell, S. | | Deposit date: | 2020-12-28 | | Release date: | 2021-01-13 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Conus venom fractions inhibit the adhesion of Plasmodium falciparum erythrocyte membrane protein 1 domains to the host vascular receptors.
J Proteomics, 234, 2020
|
|
7L83
 
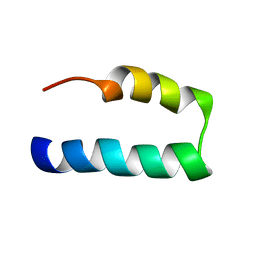 | | NMR solution structure of Nav1.5 DIV S3b-S4a paddle motif in DPC micelle | | Descriptor: | Sodium channel protein type 5 subunit alpha | | Authors: | Hussein, A.K, Bhuiyan, M.H, Arshava, B, Zhuang, J, Poget, S.F. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and analysis of isolated S3b-S4a motif of repeat IV of the human cardiac sodium channel
Biorxiv, 2021
|
|
7L8V
 
 | |
7L96
 
 | |
7L98
 
 | |
7L9D
 
 | |
7LC8
 
 | | SARS-CoV-2 spike Protein TM domain | | Descriptor: | Spike protein S2' | | Authors: | Fu, Q, Chou, J.J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Trimeric Hydrophobic Zipper Mediates the Intramembrane Assembly of SARS-CoV-2 Spike.
J.Am.Chem.Soc., 143, 2021
|
|
7LCW
 
 | | Aspartimidylated Lasso Peptide Lihuanodin | | Descriptor: | Lasso Peptide Lihuanodin | | Authors: | Link, A.J, Cao, L. | | Deposit date: | 2021-01-11 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cellulonodin-2 and Lihuanodin: Lasso Peptides with an Aspartimide Post-Translational Modification.
J.Am.Chem.Soc., 143, 2021
|
|
7LDF
 
 | |
7LGI
 
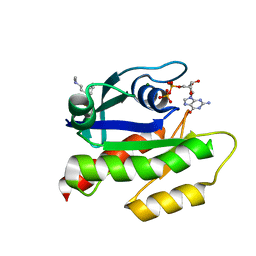 | | The haddock model of GDP KRas in complex with promazine using chemical shift perturbations and intermolecular NOEs | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, X, Gorfe, A.A, Putkey, J.A. | | Deposit date: | 2021-01-20 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Antipsychotic phenothiazine drugs bind to KRAS in vitro.
J.Biomol.Nmr, 75, 2021
|
|
7LGL
 
 | |
7LGV
 
 | |
