6IRR
 
 | | Solution structure of DISC1/ATF4 complex | | Descriptor: | Disrupted in schizophrenia 1 homolog,Cyclic AMP-dependent transcription factor ATF-4 | | Authors: | Ye, F, Yu, C, Zhang, M. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural interaction between DISC1 and ATF4 underlying transcriptional and synaptic dysregulation in an iPSC model of mental disorders.
Mol. Psychiatry, 2019
|
|
6ITH
 
 | |
6IVS
 
 | |
6IVU
 
 | |
6IWJ
 
 | | A designed domain swapped dimer | | Descriptor: | Archeal Protein MK0293 | | Authors: | Nandwani, N, Negi, H, Das, R. | | Deposit date: | 2018-12-05 | | Release date: | 2019-02-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A five-residue motif for the design of domain swapping in proteins.
Nat Commun, 10, 2019
|
|
6IWS
 
 | | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Sim, D.W, Jo, K.S, Won, H.S, Kim, J.H. | | Deposit date: | 2018-12-06 | | Release date: | 2019-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the J-domain of Tid1, a Mitochondrial Hsp40/DnaJ Protein
To Be Published
|
|
6IY5
 
 | | NMR solution structures of 5'-ATTCTATTCT-3 | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*TP*AP*TP*TP*CP*T)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Nucleic Acids Res., 48, 2020
|
|
6IYN
 
 | | Solution structure of camelid nanobody Nb26 against aflatoxin B1 | | Descriptor: | Nb26 | | Authors: | Nie, Y, He, T, Zhu, J, Li, S.L, Hu, R, Yang, Y.H. | | Deposit date: | 2018-12-17 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical shift assignments of a camelid nanobody against aflatoxin B1.
Biomol NMR Assign, 13, 2019
|
|
6IZG
 
 | |
6IZP
 
 | | Solution structure of the complex of naphthyridine carbamate dimer and an RNA with UGGAA-UGGAA pentad | | Descriptor: | 3-[3-[(7-methyl-1,8-naphthyridin-2-yl)carbamoyloxy]propylamino]propyl ~{N}-(7-methyl-1,8-naphthyridin-2-yl)carbamate, RNA (29-MER) | | Authors: | Nagano, K, Shibata, T, Nakatani, K, Kawai, G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Small molecule targeting r(UGGAA)n disrupts RNA foci and alleviates disease phenotype in Drosophila model
Nat Commun, 12, 2021
|
|
6J12
 
 | |
6J2W
 
 | | The structure of OBA3-OTA complex | | Descriptor: | DNA (5'-D(*CP*GP*GP*GP*GP*CP*GP*AP*AP*GP*CP*GP*GP*GP*TP*CP*CP*CP*G)-3'), N-[(3R)-5-chloro-8-hydroxy-3-methyl-1-oxo-3,4-dihydro-1H-2-benzopyran-7-carbonyl]-D-phenylalanine | | Authors: | Xu, G.H, Li, C.G. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure-guided post-SELEX optimization of an ochratoxin A aptamer.
Nucleic Acids Res., 47, 2019
|
|
6J2Y
 
 | |
6J37
 
 | | DNA minidumbbell structure of two CTTG repeats | | Descriptor: | DNA (5'-D(*CP*TP*TP*GP*CP*TP*TP*G)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Unprecedented hydrophobic stabilizations from a reverse wobble T·T mispair in DNA minidumbbell.
J.Biomol.Struct.Dyn., 38, 2020
|
|
6J3L
 
 | | Solution structure of the N-terminal extended protuberant domain of eukaryotic ribosomal stalk protein P0 | | Descriptor: | 60S acidic ribosomal protein P0 | | Authors: | Choi, K.H.A, Lee, K.M, Yang, L, Wing-Heng Yu, C, Banfield, D.K, Ito, K, Uchiumi, T, Wong, K.B. | | Deposit date: | 2019-01-04 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Mutagenesis Studies Evince the Role of the Extended Protuberant Domain of Ribosomal Protein uL10 in Protein Translation.
Biochemistry, 58, 2019
|
|
6J4I
 
 | |
6J9P
 
 | |
6JCD
 
 | |
6JCE
 
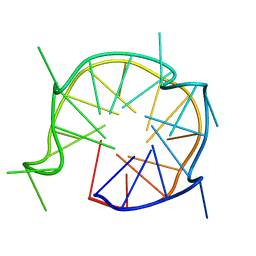 | | NMR solution and X-ray crystal structures of a DNA containing both right-and left-handed parallel-stranded G-quadruplexes | | Descriptor: | 29-mer DNA | | Authors: | Winnerdy, F.R, Bakalar, B, Maity, A, Vandana, J.J, Mechulam, Y, Schmitt, E, Phan, A.T. | | Deposit date: | 2019-01-28 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution and X-ray crystal structures of a DNA molecule containing both right- and left-handed parallel-stranded G-quadruplexes.
Nucleic Acids Res., 47, 2019
|
|
6JHD
 
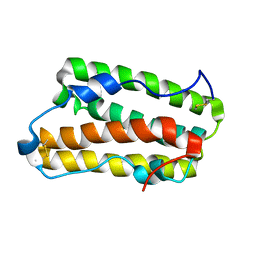 | | Solution structure of IFN alpha8 | | Descriptor: | Interferon alpha-8 | | Authors: | Ken-ichi, A, Shigeyuki, M. | | Deposit date: | 2019-02-18 | | Release date: | 2020-02-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Determination of solution structure of interferon alpha8:
Novel insights into the preferred interaction with IFNAR2 among its subtypes
To Be Published
|
|
6JI7
 
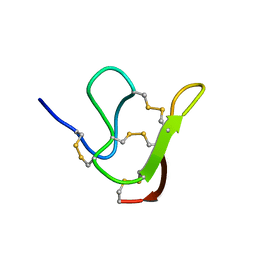 | |
6JIC
 
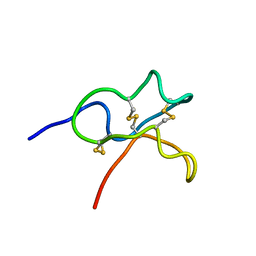 | | Identification and Characterization of a carboxypeptidase inhibitor from Lycium barbarum | | Descriptor: | WCI | | Authors: | Tan, W.L, Wong, K.H, Huang, J.Y, Tay, S.V, Wang, S.J. | | Deposit date: | 2019-02-20 | | Release date: | 2020-02-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and characterization of a wolfberry carboxypeptidase inhibitor from Lycium barbarum.
Food Chem, 351, 2021
|
|
6JJ0
 
 | |
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | Deposit date: | 2019-03-26 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
6JPP
 
 | |
