8QZK
 
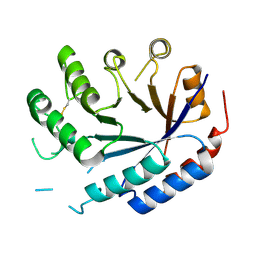 | | Catalytic core of endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum (EngBF) concieved by deep network hallucination: dEngBF4 Hexagonal form | | Descriptor: | ENDO-ALPHA-N-ACETYLGALACTOSAMINIDASE | | Authors: | Aghajari, N, Hansen, A.L, Thiesen, F.F, Crehuet, R, Marcos, E, Willemoes, M. | | Deposit date: | 2023-10-27 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Carving out a Glycoside Hydrolase Active Site for Incorporation into a New Protein Scaffold Using Deep Network Hallucination.
Acs Synth Biol, 13, 2024
|
|
8QZS
 
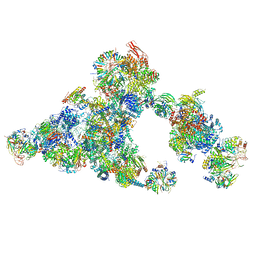 | | Cryo-EM structure of the cross-exon B-like complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Microfibrillar-associated protein 1, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-29 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R08
 
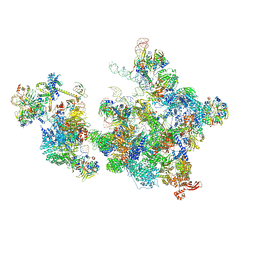 | | Cryo-EM structure of the cross-exon pre-B+AMPPNP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, NHP2-like protein 1, N-terminally processed, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R09
 
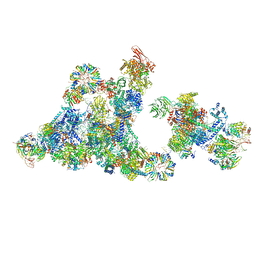 | | Cryo-EM structure of the cross-exon pre-B+5'ss+ATPgammaS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R0A
 
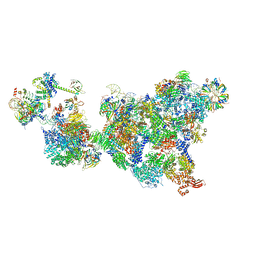 | | Cryo-EM structure of the cross-exon pre-B+5'ss complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R0B
 
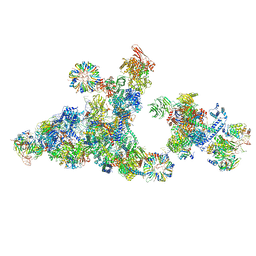 | | Cryo-EM structure of the cross-exon pre-B+ATP complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'ss oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2023-10-31 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
8R3V
 
 | |
8R55
 
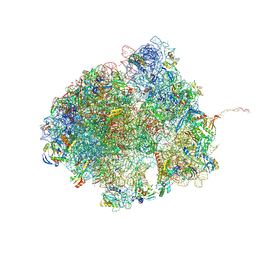 | | Bacillus subtilis MutS2-collided disome complex (collided 70S) | | Descriptor: | 16S rRNA (1533-MER), 23S RNA (2887-MER), 30S ribosomal protein S10, ... | | Authors: | Park, E, Mackens-Kiani, T, Berhane, R, Esser, H, Erdenebat, C, Burroughs, A.M, Berninghausen, O, Aravind, L, Beckmann, R, Green, R, Buskirk, A.R. | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | B. subtilis MutS2 splits stalled ribosomes into subunits without mRNA cleavage.
Embo J., 43, 2024
|
|
8R5O
 
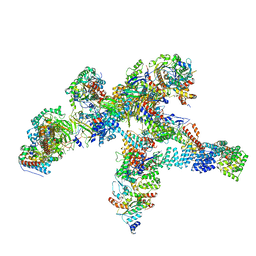 | | Plastid-encoded RNA polymerase | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-17 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8R6C
 
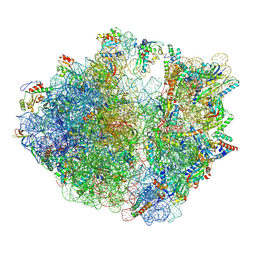 | |
8R6G
 
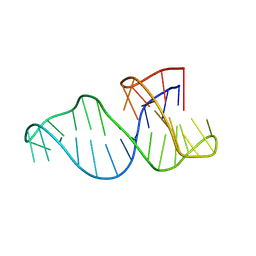 | |
8R6S
 
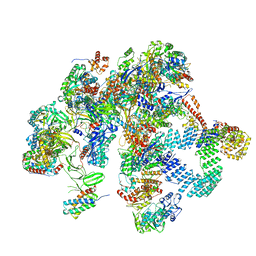 | | Plastid-encoded RNA polymerase (Integrated model) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-11-22 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8R8M
 
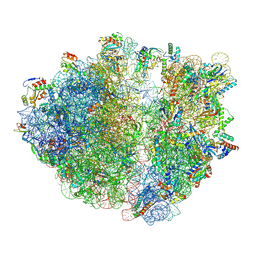 | |
8RAS
 
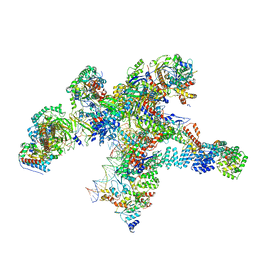 | | Plastid-encoded RNA polymerase transcription elongation complex | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8RBS
 
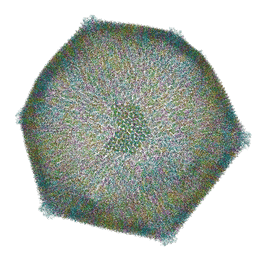 | | Emiliania huxleyi virus 201 (EhV-201) asymmetrical unit of capsid proteins predicted by AlphaFold2 fitted into the cryo-EM density of EhV-201 virion composite map. | | Descriptor: | Major capsid protein, Penton protein | | Authors: | Homola, M, Buttner, C.R, Fuzik, T, Novacek, J, Chaillet, M, Forster, F, Plevka, P. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure and replication cycle of a virus infecting climate-modulating alga Emiliania huxleyi.
Sci Adv, 10, 2024
|
|
8RBX
 
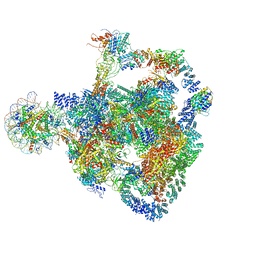 | | Structure of Integrator-PP2A bound to a paused RNA polymerase II-DSIF-NELF-nucleosome complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit RPB11-a, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Fianu, I, Ochmann, M, Walshe, J.L, Cramer, P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of Integrator-dependent RNA polymerase II termination.
Nature, 629, 2024
|
|
8RCL
 
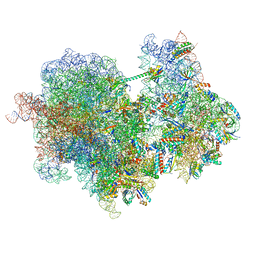 | |
8RCM
 
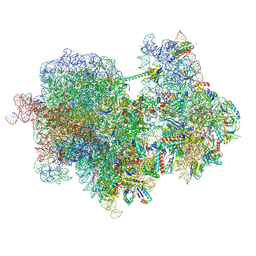 | |
8RCS
 
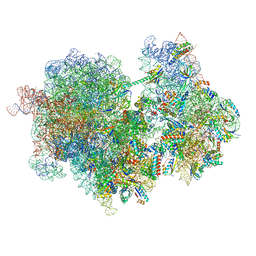 | |
8RCT
 
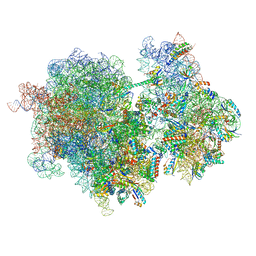 | |
8RD8
 
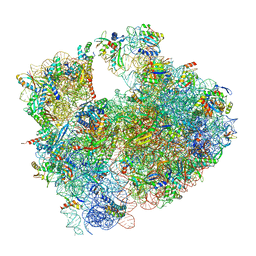 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factors Balon and RaiA (structure 1). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S30, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-07 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8RDA
 
 | |
8RDF
 
 | |
8RDJ
 
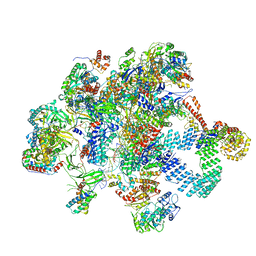 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8RDP
 
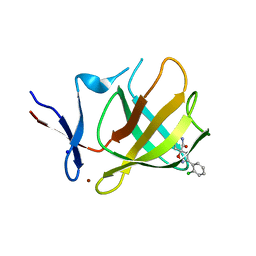 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with spiro-isoxazol based compound 8a | | Descriptor: | (5~{S})-3-(2-chlorophenyl)-1-oxa-2,9-diazaspiro[4.5]dec-2-ene-8,10-dione, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Discovery and characterization of potent spiro-isoxazole-based cereblon ligands with a novel binding mode.
Eur.J.Med.Chem., 270, 2024
|
|






