8D9U
 
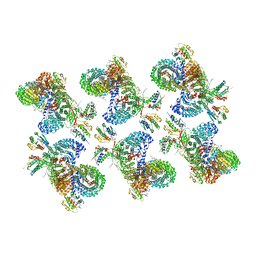 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on beta-Arf1 | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9V
 
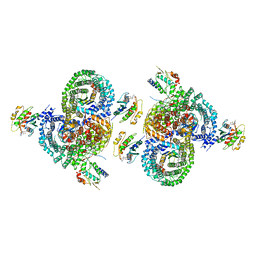 | | gamma-Arf1 homodimeric interface within AP-1, Arf1, Nef lattice on narrow membrane tubes | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8D9W
 
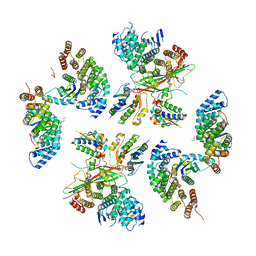 | | beta-Arf1 homodimeric interface within AP-1, Arf1, Nef, MHC-I lattice on narrow tubes | | Descriptor: | ADP-ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.H, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
8DAP
 
 | | [GA/TC] Self-Assembled 3D DNA Tensegrity Triangle with 24 bp Arm Length forming a Trigonal Hexagon | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*CP*TP*AP*CP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*GP*AP*TP*GP*TP*GP*GP*CP*TP*AP*GP*GP*C)-3'), DNA (5'-D(P*TP*AP*CP*AP*CP*CP*GP*TP*AP*CP*AP*CP*CP*GP*TP*AP*CP*AP*CP*CP*G)-3') | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-06-13 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.47 Å) | | Cite: | Highly Symmetric, Self-Assembling 3D DNA Crystals with Cubic and Trigonal Lattices.
Small, 19, 2023
|
|
8DBP
 
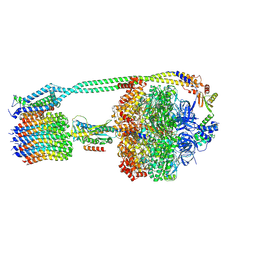 | | E. coli ATP synthase imaged in 10mM MgATP State1 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBQ
 
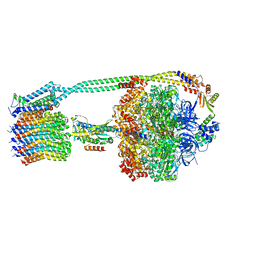 | |
8DBR
 
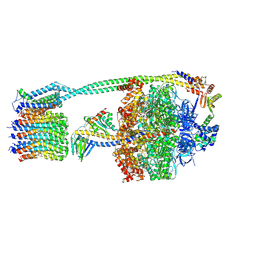 | | E. coli ATP synthase imaged in 10mM MgATP State2 "half-up | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBT
 
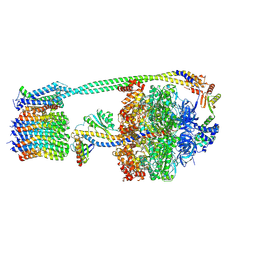 | | E. coli ATP synthase imaged in 10mM MgATP State2 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBU
 
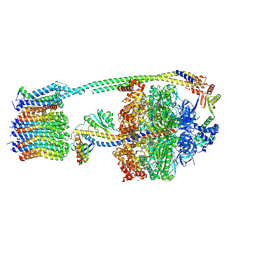 | |
8DBV
 
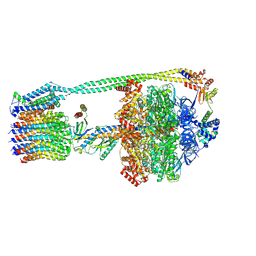 | | E. coli ATP synthase imaged in 10mM MgATP State3 "down | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Sobti, M, Stewart, A.G. | | Deposit date: | 2022-06-14 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Changes within the central stalk of E. coli F 1 F o ATP synthase observed after addition of ATP.
Commun Biol, 6, 2023
|
|
8DBW
 
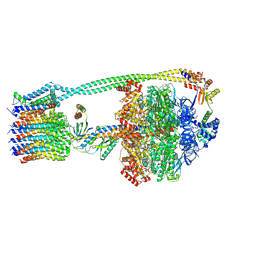 | |
8DCM
 
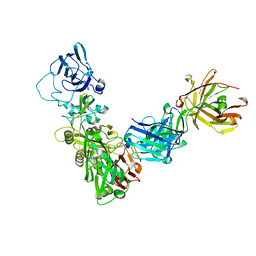 | |
8DD3
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
8DE6
 
 | | Oligomeric C9 in complex with aE11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement component C9, ... | | Authors: | Bayly-Jones, C. | | Deposit date: | 2022-06-20 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The neoepitope of the complement C5b-9 Membrane Attack Complex is formed by proximity of adjacent ancillary regions of C9.
Commun Biol, 6, 2023
|
|
8DHX
 
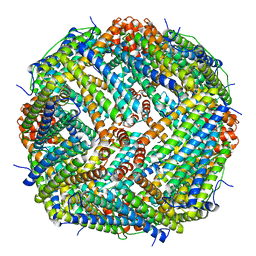 | | Human liver ferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Zhang, Z. | | Deposit date: | 2022-06-28 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Human liver ferritin
To Be Published
|
|
8DLI
 
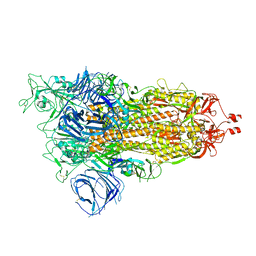 | | Cryo-EM structure of SARS-CoV-2 Alpha (B.1.1.7) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLT
 
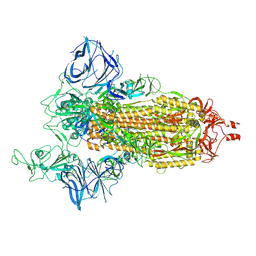 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLU
 
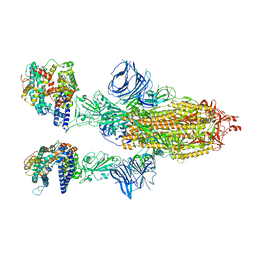 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLX
 
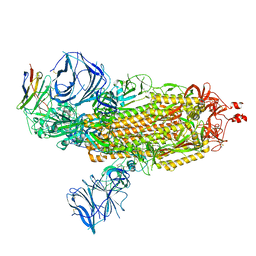 | | Cryo-EM structure of SARS-CoV-2 Epsilon (B.1.429) spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DLZ
 
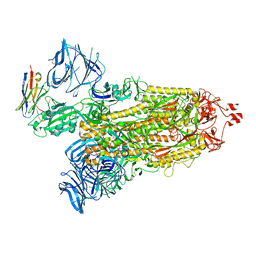 | | Cryo-EM structure of SARS-CoV-2 D614G spike protein in complex with VH ab6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization.
Nat Commun, 13, 2022
|
|
8DN3
 
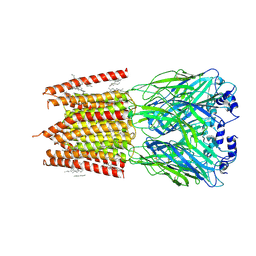 | |
8DNT
 
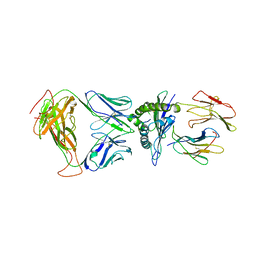 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
8DP1
 
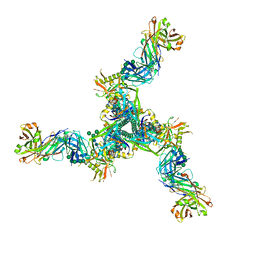 | | Cryo-EM structure of HIV-1 Env(BG505.T332N SOSIP) in complex with DH1030.1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DH1030.1 Fab Heavy chain, ... | | Authors: | Gobeil, S, Acharya, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Shared recognition mechanism for HIV-1 envelope-reactive V3 glycan broadly neutralizing B cell lineage maturation in humans and macaques
To Be Published
|
|
8DP8
 
 | |
8DP9
 
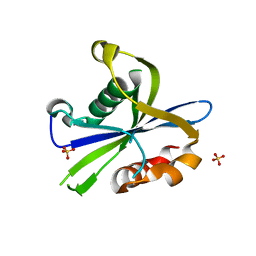 | | Crystal structure of the monomeric AvrM14-B Nudix hydrolase effector from Melampsora lini | | Descriptor: | AvrM14-B, SULFATE ION | | Authors: | McCombe, C.L, Outram, M.A, Ericsson, D.J, Williams, S.J. | | Deposit date: | 2022-07-15 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A rust-fungus Nudix hydrolase effector decaps mRNA in vitro and interferes with plant immune pathways.
New Phytol., 239, 2023
|
|






