9C0B
 
 | | E.coli GroEL + PBZ1587 inhibitor | | Descriptor: | 60 kDa chaperonin, N-(2-{4-[4-(aminomethyl)benzene-1-sulfonamido]phenyl}-1,3-benzoxazol-5-yl)-5-chlorothiophene-2-sulfonamide | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2024-05-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Bis-sulfonamido-2-phenylbenzoxazoles Validate the GroES/EL Chaperone System as a Viable Antibiotic Target.
J.Am.Chem.Soc., 146, 2024
|
|
9C0Y
 
 | | Clathrin terminal domain complexed with Pitstop 2c | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Clathrin heavy chain 1, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
9C0Z
 
 | | Clathrin terminal domain complexed with pitstop 2d | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Bulut, H, Horatscheck, A, Krauss, M, Santos, K.F, McCluskey, A, Wahl, C.W, Nazare, M, Haucke, V. | | Deposit date: | 2024-05-28 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Acute inhibition of clathrin-mediated endocytosis by next-generation small molecule inhibitors of clathrin function
To Be Published
|
|
9C10
 
 | | AMG 193, a clinical stage MTA-cooperative PRMT5 inhibitor, drives anti-tumor activity preclinically and in patients with MTAP-deleted cancers | | Descriptor: | (4-amino-1,3-dihydrofuro[3,4-c][1,7]naphthyridin-8-yl){(3S)-3-[4-(trifluoromethyl)phenyl]morpholin-4-yl}methanone, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DIMETHYL SULFOXIDE, ... | | Authors: | Ghimire-Rijal, S, Mukund, S. | | Deposit date: | 2024-05-28 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | AMG 193, a Clinical Stage MTA-Cooperative PRMT5 Inhibitor, Drives Antitumor Activity Preclinically and in Patients With MTAP-Deleted Cancers.
Cancer Discov, 2024
|
|
9C1P
 
 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '6218 | | Descriptor: | (5R)-N-[2-(1,2-benzothiazol-3-yl)ethyl]-1-methyl-2,3,4,5-tetrahydro-1H-1-benzazepin-5-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-29 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9C1Q
 
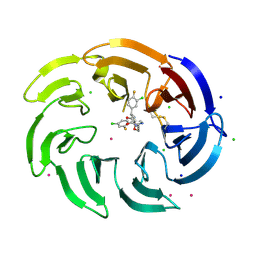 | | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-39512 compound | | Descriptor: | (4P)-N-[(1S)-3-amino-1-(3-chloro-4-fluorophenyl)-3-oxopropyl]-4-(4-chloro-2-fluorophenyl)-5-(3-methoxyprop-1-yn-1-yl)-1H-pyrrole-3-carboxamide, CHLORIDE ION, DDB1- and CUL4-associated factor 1, ... | | Authors: | kimani, S, Dong, A, Li, Y, Seitova, A, Al-awar, R, Wilson, B, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-05-29 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the WDR domain of human DCAF1 in complex with OICR-39512 compound
To be published
|
|
9C1R
 
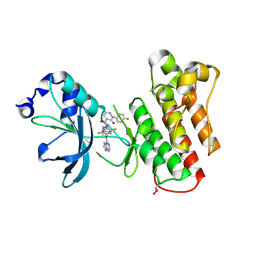 | | Crystal structure of mutant cMET D1228N kinase domain in complex with inhibitor compound 13 | | Descriptor: | GLYCEROL, Hepatocyte growth factor receptor, N-(2,5-difluoro-4-{[(1s,3S)-3-(1-methyl-1H-pyrazol-3-yl)cyclobutyl][(8R)-pyrazolo[1,5-a]pyrazin-4-yl]amino}phenyl)-2-(5-fluoropyridin-2-yl)-3-oxo-2,3-dihydropyridazine-4-carboxamide | | Authors: | Simpson, H, Wu, W.-I, Mou, T.-C. | | Deposit date: | 2024-05-29 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Discovery of Pyrazolopyrazines as Selective, Potent, and Mutant-Active MET Inhibitors with Intracranial Efficacy.
J.Med.Chem., 67, 2024
|
|
9C2A
 
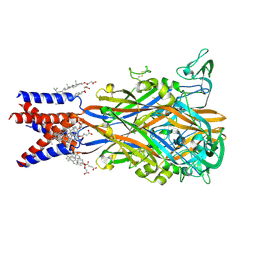 | | Cryo-EM structure of the human P2X1 receptor in the apo closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
9C2B
 
 | | Cryo-EM structure of the human P2X1 receptor in the ATP-bound desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
9C2C
 
 | | Cryo-EM structure of the human P2X1 receptor in the NF449-bound inhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4',4'',4'''-{carbonylbis[azanediylbenzene-5,1,3-triylbis(carbonylazanediyl)]}tetra(benzene-1,3-disulfonic acid), P2X purinoceptor 1 | | Authors: | Oken, A.C, Lisi, N.E, Ditter, I.A, Shi, H, Mansoor, S.E. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structures of the human P2X1 receptor reveal subtype-specific architecture and antagonism by supramolecular ligand-binding.
Nat Commun, 15, 2024
|
|
9C2F
 
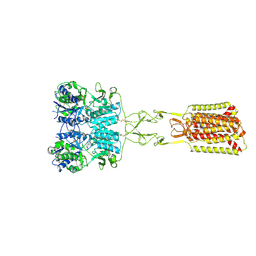 | | Structure of Calcium-Sensing Receptor in complex with positive allosteric modulator '54149 | | Descriptor: | (1R)-1-(2H-1,3-benzodioxol-4-yl)-N-[2-(1,2-benzothiazol-3-yl)ethyl]ethan-1-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, C, Skiniotis, G. | | Deposit date: | 2024-05-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Large library docking identifies positive allosteric modulators of the calcium-sensing receptor.
Science, 385, 2024
|
|
9C4I
 
 | | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Nascimento, K.S, Pinto-Junior, V.R, Lima, F.E.O, Osterne, V.J.S, Oliveira, M.V, Ferreira, V.M.S, Cavada, B.S. | | Deposit date: | 2024-06-04 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Centrolobium microchaete seed lectin (CML) complexed with Man1-3Man-OMe
To Be Published
|
|
9C4M
 
 | | Crystal Structure of A. baumannii GuaB dCBS with inhibitor G6 (3826) | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, N-[4-chloro-3-(morpholin-4-yl)phenyl]-N~2~-[3-(hydroxymethyl)quinolin-6-yl]-L-alaninamide | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2024-06-04 | | Release date: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Differential effects of inosine monophosphate dehydrogenase (IMPDH/GuaB) inhibition in Acinetobacter baumannii and Escherichia coli.
J.Bacteriol., 2024
|
|
9C5D
 
 | | E. coli peptidyl-prolyl cis-trans isomerase containing (2S,3S)-4-Fluorovaline | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ACETATE ION, ... | | Authors: | Frkic, R.L, Jackson, C.J. | | Deposit date: | 2024-06-06 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | 1.3 angstrom Crystal Structure of E. coli Peptidyl-Prolyl Isomerase B with Uniform Substitution of Valine by (2 S ,3 S )-4-Fluorovaline Reveals Structure Conservation and Multiple Staggered Rotamers of CH 2 F Groups.
Biochemistry, 2024
|
|
9C61
 
 | | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26 | | Descriptor: | (4R)-4-[4-(5-fluoro-1H-indol-3-yl)piperidine-1-carbonyl]piperidin-2-one, Non-specific serine/threonine protein kinase, UNKNOWN ATOM OR ION | | Authors: | Zeng, H, Dong, A, Kutera, M, Ilyassov, O, Seitova, A, Loppnau, P, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-06-07 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the human LRRK2 WDR domain in complex with CACHE1193-26
To be published
|
|
9C62
 
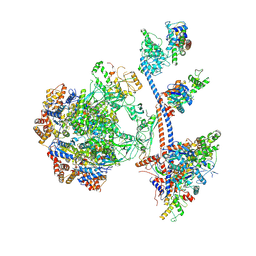 | | P400 subcomplex of the native human TIP60 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Yang, Z, Mameri, A, Florez Ariza, A.J, Cote, J, Nogales, E. | | Deposit date: | 2024-06-07 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (5.28 Å) | | Cite: | Structural insights into the human NuA4/TIP60 acetyltransferase and chromatin remodeling complex.
Science, 385, 2024
|
|
9C6P
 
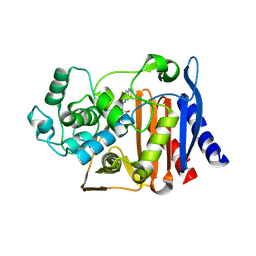 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | 3-chloro-N-(5-chloro-2-methyl-1,3-benzothiazol-6-yl)-2-hydroxybenzene-1-sulfonamide, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-08 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.663 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C7V
 
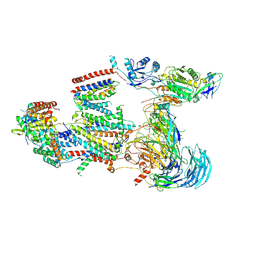 | | Structure of the human BOS:human EMC complex in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BOS complex subunit NOMO2, ... | | Authors: | Nguyen, V.N, Tomaleri, G.P, Voorhees, R.M. | | Deposit date: | 2024-06-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Role of a holo-insertase complex in the biogenesis of biophysically diverse ER membrane proteins.
Mol.Cell, 84, 2024
|
|
9C81
 
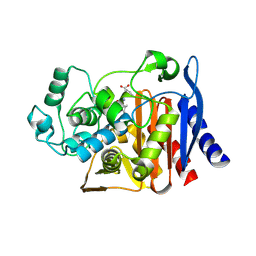 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | (2R)-2-phenoxy-3-{[(1S,2S,4S)-spiro[bicyclo[2.2.1]heptane-7,1'-cyclopropane]-2-carbonyl]amino}propanoic acid, AmpC Beta-lactamase | | Authors: | Liu, F, Shoichet, B.K, Bassim, V. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C82
 
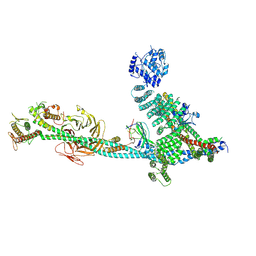 | | Structure of human ULK1C:PI3KC3-C1 supercomplex | | Descriptor: | Beclin 1-associated autophagy-related key regulator, Beclin-1, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2024-06-11 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.84 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
9C83
 
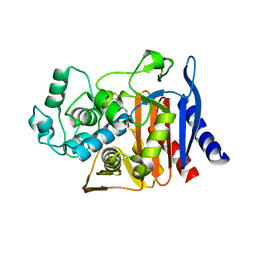 | | X-ray crystal structure of AmpC beta-lactamase with inhibitor | | Descriptor: | AmpC Beta-lactamase, N-[(3M)-3-(5-chloro-1,2,3-thiadiazol-4-yl)phenyl]-5-methyl-3-oxo-2,3-dihydro-1,2-oxazole-4-sulfonamide | | Authors: | Liu, F, Shoichet, B.K. | | Deposit date: | 2024-06-11 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Improved correlations with score, hit-rate, and affinity as docking library and testing scale increase
To Be Published
|
|
9C84
 
 | |
9C8J
 
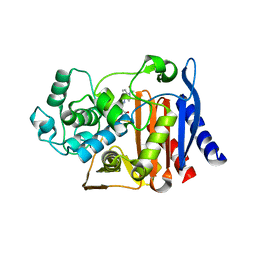 | |
9C9V
 
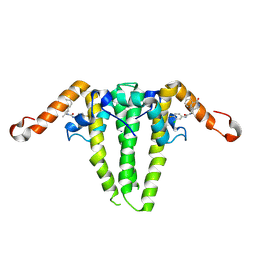 | | HBV capsid with compound 3i | | Descriptor: | Capsid protein, N'-(3-chloro-4-fluorophenyl)-N-(2-methylpropyl)-N-[(1R)-1-(1-oxo-1,2-dihydroisoquinolin-4-yl)ethyl]urea | | Authors: | Olland, A.M, Suto, R.K, Fontano, E, Colussi, T. | | Deposit date: | 2024-06-16 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Rational Design, Synthesis, and Structure-Activity Relationship of a Novel Isoquinolinone-Based Series of HBV Capsid Assembly Modulators Leading to the Identification of Clinical Candidate AB-836.
J.Med.Chem., 67, 2024
|
|
9CAI
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | High-Resolution Reconstruction of a C. elegans Ribosome Sheds Light on Evolutionary Dynamics and Tissue Specificity.
Rna, 2024
|
|






