6Y69
 
 | |
6Y6X
 
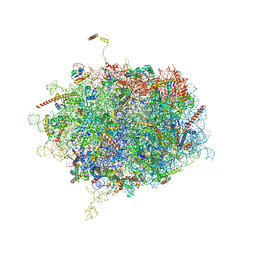 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
6Y7M
 
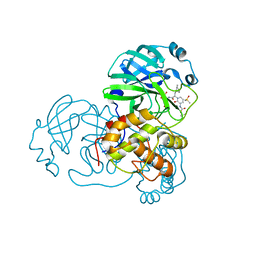 | | Crystal structure of the complex resulting from the reaction between the SARS-CoV main protease and tert-butyl (1-((S)-3-cyclohexyl-1-(((S)-4-(cyclopropylamino)-3,4-dioxo-1-((S)-2-oxopyrrolidin-3-yl)butan-2-yl)amino)-1-oxopropan-2-yl)-2-oxo-1,2-dihydropyridin-3-yl)carbamate | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, ~{tert}-butyl ~{N}-[1-[(2~{S})-3-cyclohexyl-1-[[(2~{S},3~{R})-4-(cyclopropylamino)-3-oxidanyl-4-oxidanylidene-1-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butan-2-yl]amino]-1-oxidanylidene-propan-2-yl]-2-oxidanylidene-pyridin-3-yl]carbamate | | Authors: | Zhang, L, Lin, D, Hilgenfeld, R. | | Deposit date: | 2020-03-01 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors.
Science, 368, 2020
|
|
6Y7Q
 
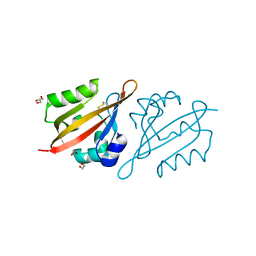 | | Crystal Structure of the N-terminal PAS domain from the hERG3 Potassium Channel | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Potassium voltage-gated channel subfamily H member 7 | | Authors: | Cresser-Brown, J, Raven, E, Moody, P. | | Deposit date: | 2020-03-02 | | Release date: | 2020-08-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of a heme-binding domain in a neuronal voltage-gated potassium channel.
J.Biol.Chem., 295, 2020
|
|
6Y8G
 
 | | selenomethionine derivative of ferulic acid esterase (FAE) | | Descriptor: | CADMIUM ION, Endo-1,4-beta-xylanase Y, GLYCEROL | | Authors: | von Stetten, D, Mueller-Dieckmann, C, Carpentier, P, Flot, D. | | Deposit date: | 2020-03-04 | | Release date: | 2020-05-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ID30A-3 (MASSIF-3) - a beamline for macromolecular crystallography at the ESRF with a small intense beam.
J.Synchrotron Radiat., 27, 2020
|
|
6Y8K
 
 | | Crystal structure of CD137 in complex with the cyclic peptide BCY10916 | | Descriptor: | 1,1',1''-(1,3,5-triazinane-1,3,5-triyl)tripropan-1-one, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Upadhyaya, P, Kublin, J, Dods, R, Kristensson, J, Lahdenranta, J, Kleyman, M, Repash, E, Ma, J, Mudd, G, Van Rietschoten, K, Haines, E, Harrison, H, Beswick, P, Chen, L, McDonnell, K, Battula, S, Hurov, K, Keen, N. | | Deposit date: | 2020-03-05 | | Release date: | 2021-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Anticancer immunity induced by a synthetic tumor-targeted CD137 agonist.
J Immunother Cancer, 9, 2021
|
|
6Y8X
 
 | |
6Y9I
 
 | |
6Y9K
 
 | |
6YAB
 
 | |
6YAJ
 
 | | Split gene transketolase, inactive beta4 tetramer | | Descriptor: | 1,2-ETHANEDIOL, C-terminal chain of split transketolase from Carboxydothermus hydrogenoformans, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
6YAK
 
 | | Split gene transketolase, active alpha2beta2 heterotetramer | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-2H-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, C-terminal component of the split chain transketolase, ... | | Authors: | Isupov, M.N, Littlechild, J.A, James, P. | | Deposit date: | 2020-03-12 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A 'Split-Gene' Transketolase From the Hyper-Thermophilic Bacterium Carboxydothermus hydrogenoformans : Structure and Biochemical Characterization.
Front Microbiol, 11, 2020
|
|
6YAM
 
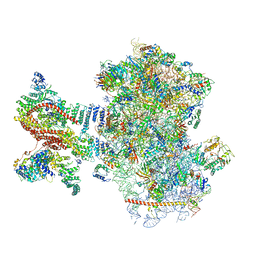 | | Mammalian 48S late-stage translation initiation complex (LS48S+eIF3 IC) with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein eS1, 40S ribosomal protein eS10, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6YAX
 
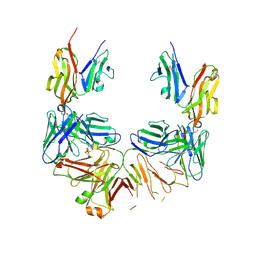 | |
6YB4
 
 | |
6YD2
 
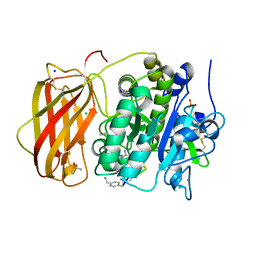 | |
6YD3
 
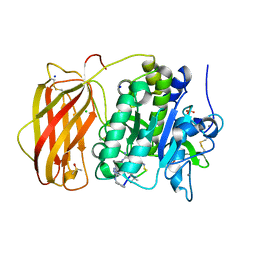 | |
6YDP
 
 | |
6YDV
 
 | | Crystal Structure of the Jmjc Domain of Human JMJD1B in complex with FM001511a from the DSPL fragment library | | Descriptor: | CHLORIDE ION, JMJD1B protein, MANGANESE (II) ION, ... | | Authors: | Snee, M, Nowak, R, Johansson, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of the Jmjc Domain of Human JMJD1B in complex with FM001511a from the DSPL fragment library
To Be Published
|
|
6YDW
 
 | |
6YEF
 
 | | 70S initiation complex with assigned rRNA modifications from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Fatkhullin, B, Golubev, A, Khusainov, I, Yusupova, G, Yusupov, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-27 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the ribosome functional complex of the human pathogen Staphylococcus aureus at 3.2 angstrom resolution.
Febs Lett., 594, 2020
|
|
6YF7
 
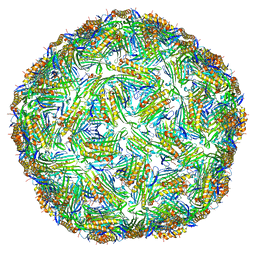 | | Virus-like particle of bacteriophage AC | | Descriptor: | Coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YF9
 
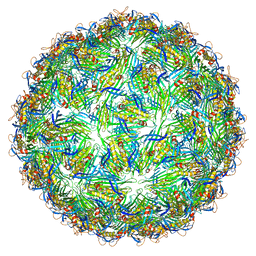 | | Virus-like particle of bacteriophage AVE002 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.196 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFA
 
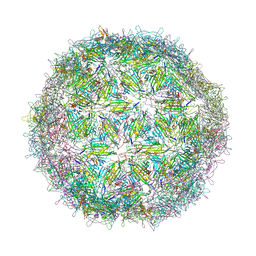 | | Virus-like particle of bacteriophage AVE015 | | Descriptor: | CALCIUM ION, coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|
6YFB
 
 | | Virus-like particle of bacteriophage AVE016 | | Descriptor: | coat protein | | Authors: | Rumnieks, J, Kalnins, G, Sisovs, M, Lieknina, I, Tars, K. | | Deposit date: | 2020-03-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes.
Sci Adv, 6, 2020
|
|







