6QCF
 
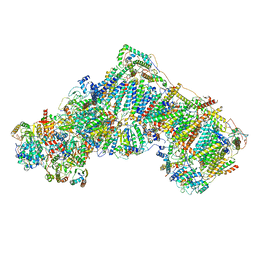 | | Ovine respiratory complex I FRC open class 6 | | Descriptor: | Acyl carrier protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Letts, J.A, Sazanov, L.A. | | Deposit date: | 2018-12-27 | | Release date: | 2019-08-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structures of Respiratory Supercomplex I+III2Reveal Functional and Conformational Crosstalk.
Mol.Cell, 75, 2019
|
|
6QCM
 
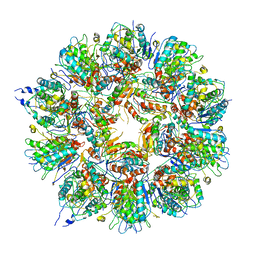 | | Cryo em structure of the Listeria stressosome | | Descriptor: | RsbR protein, RsbR protein,RsbR protein, RsbS protein | | Authors: | Williams, A.H, Redzej, A, Waksman, G, Cossart, P. | | Deposit date: | 2018-12-28 | | Release date: | 2019-08-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The cryo-electron microscopy supramolecular structure of the bacterial stressosome unveils its mechanism of activation.
Nat Commun, 10, 2019
|
|
6QDV
 
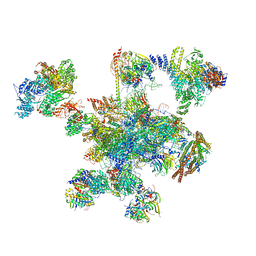 | | Human post-catalytic P complex spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent RNA helicase DHX8, ... | | Authors: | Fica, S.M, Oubridge, C, Wilkinson, M.E, Newman, A.J, Nagai, K. | | Deposit date: | 2019-01-03 | | Release date: | 2019-02-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation.
Science, 363, 2019
|
|
6QI5
 
 | | Near Atomic Structure of an Atadenovirus Shows a possible gene duplication event and Intergenera Variations in Cementing Proteins | | Descriptor: | Hexon protein, PIIIa, Penton protein, ... | | Authors: | Condezo, G.N, Marabini, R, Gomez-Blanco, J, SanMartin, C. | | Deposit date: | 2019-01-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-atomic structure of an atadenovirus reveals a conserved capsid-binding motif and intergenera variations in cementing proteins.
Sci Adv, 7, 2021
|
|
6QIK
 
 | |
6QL5
 
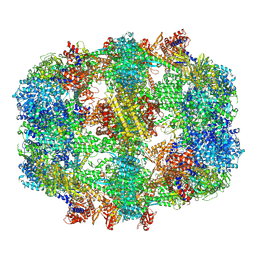 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 2.8 angstrom | | Descriptor: | 4'-PHOSPHOPANTETHEINE, FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL6
 
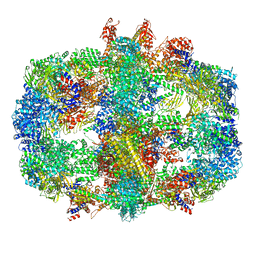 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL7
 
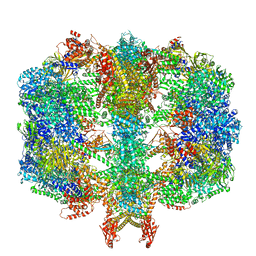 | | Structure of fatty acid synthase complex with bound gamma subunit from Saccharomyces cerevisiae at 4.6 angstrom | | Descriptor: | Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, Translation machinery-associated protein 17 | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QL9
 
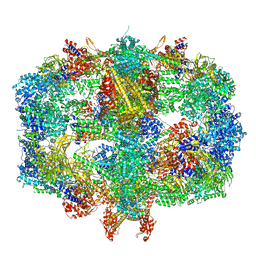 | | Structure of Fatty acid synthase complex from Saccharomyces cerevisiae at 2.9 Angstrom | | Descriptor: | 1,2-ETHANEDIOL, 4'-PHOSPHOPANTETHEINE, ACETATE ION, ... | | Authors: | Singh, K, Graf, B, Linden, A, Sautner, V, Urlaub, H, Tittmann, K, Stark, H, Chari, A. | | Deposit date: | 2019-01-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery of a Regulatory Subunit of the Yeast Fatty Acid Synthase.
Cell, 180, 2020
|
|
6QN1
 
 | | T=4 quasi-symmetric bacterial microcompartment particle | | Descriptor: | BMC domain-containing protein, Carbon dioxide concentrating mechanism protein CcmL | | Authors: | Kalnins, G. | | Deposit date: | 2019-02-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles.
Nat Commun, 11, 2020
|
|
6QNQ
 
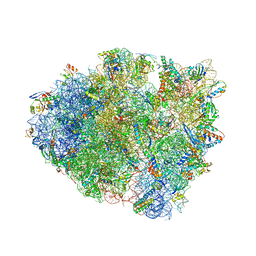 | | 70S ribosome initiation complex (IC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
6QNR
 
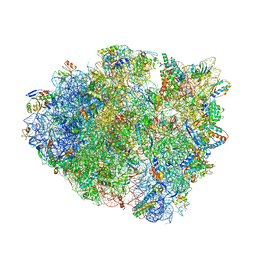 | | 70S ribosome elongation complex (EC) with experimentally assigned potassium ions | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2019-02-11 | | Release date: | 2019-06-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction.
Nat Commun, 10, 2019
|
|
6QSW
 
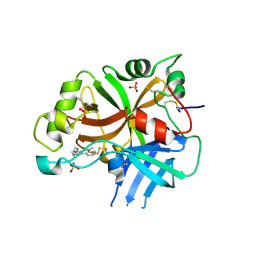 | | Complement factor B protease domain in complex with the reversible inhibitor N-(2-bromo-4-methylnaphthalen-1-yl)-4,5-dihydro-1H-imidazol-2-amine. | | Descriptor: | Complement factor B, SULFATE ION, ~{N}-(2-bromanyl-4-methyl-naphthalen-1-yl)-4,5-dihydro-1~{H}-imidazol-2-amine | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QSX
 
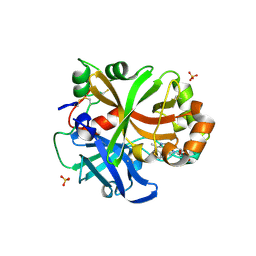 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6QT0
 
 | |
6QTZ
 
 | |
6QVK
 
 | |
6QW6
 
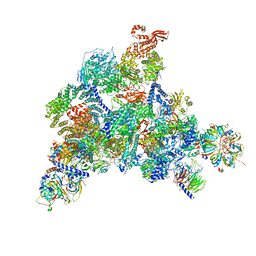 | | Structure of the human U5.U4/U6 tri-snRNP at 2.9A resolution. | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-05 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
6QX7
 
 | |
6QX9
 
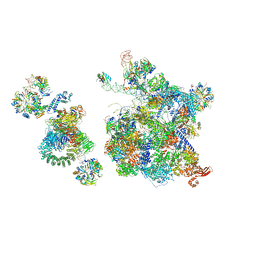 | | Structure of a human fully-assembled precatalytic spliceosome (pre-B complex). | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, AdML pre-mRNA, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Charenton, C, Wilkinson, M.E, Nagai, K. | | Deposit date: | 2019-03-07 | | Release date: | 2019-04-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanism of 5' splice site transfer for human spliceosome activation.
Science, 364, 2019
|
|
6QYD
 
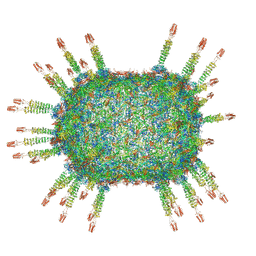 | | Cryo-EM structure of the head in mature bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J.W, Wang, D.H, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
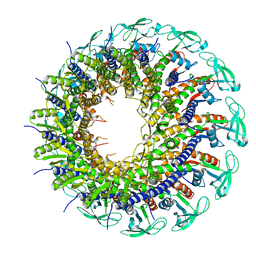
6QYJ
 
 | |
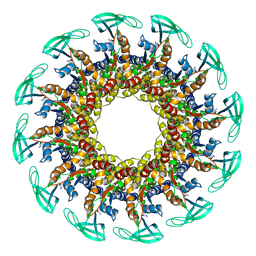
6QYM
 
 | |
6QZ0
 
 | | The cryo-EM structure of the head of the genome empited bacteriophage phi29 | | Descriptor: | Capsid fiber protein, Major capsid protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|
6QZ9
 
 | | The cryo-EM structure of the collar complex and tail axis in bacteriophage phi29 | | Descriptor: | Portal protein, Pre-neck appendage protein, Proximal tail tube connector protein | | Authors: | Xu, J, Wang, D, Gui, M, Xiang, Y. | | Deposit date: | 2019-03-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural assembly of the tailed bacteriophage φ29.
Nat Commun, 10, 2019
|
|







