9C8J
 
 | |
9CAI
 
 | | High-resolution C. elegans 80S ribosome structure - class 2 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-06-17 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | High-Resolution Reconstruction of a C. elegans Ribosome Sheds Light on Evolutionary Dynamics and Tissue Specificity.
Rna, 2024
|
|
9CBF
 
 | |
9CBG
 
 | |
9CBH
 
 | |
9CBI
 
 | |
9CBJ
 
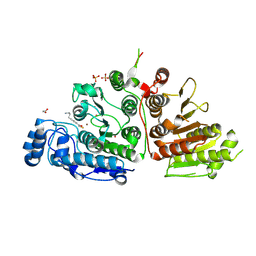 | |
9CBK
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 6 in Complex with p-Aminomethyl Phenylthioketone | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(aminomethyl)phenyl]-2-sulfanylethan-1-one, Hdac6 protein, ... | | Authors: | Goulart Stollmaier, J, Watson, P.R, Christianson, D.W. | | Deposit date: | 2024-06-19 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Design, Synthesis, and Structural Evaluation of Acetylated Phenylthioketone Inhibitors of HDAC10
To be Published
|
|
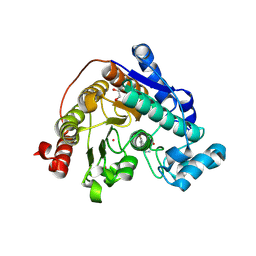
9CC9
 
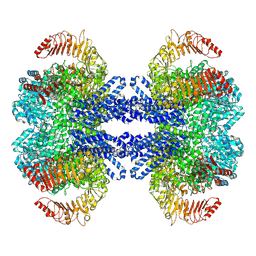 | |
9CGM
 
 | | The Structure of Spiroplasma Virus 4 | | Descriptor: | Capsid protein VP1, DNA binding protein ORF8 | | Authors: | Mietzsch, M, McKenna, R. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | The Structure of Spiroplasma Virus 4 : Exploring the Capsid Diversity of the Microviridae.
Viruses, 16, 2024
|
|
9CIW
 
 | | Penguinpox cGAMP PDE H72A mutant in complex with 2'3'-cGAMP | | Descriptor: | Penguinpox cGAMP PDE, [(2~{R},3~{R},4~{R},5~{S})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl [(2~{R},3~{R},4~{S},5~{R})-2-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl] hydrogen phosphate | | Authors: | Hobbs, S.J, Nomburg, J, Doudna, J.A, Kranzusch, P.J. | | Deposit date: | 2024-07-05 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Animal and bacterial viruses share conserved mechanisms of immune evasion.
Cell, 2024
|
|
9CL9
 
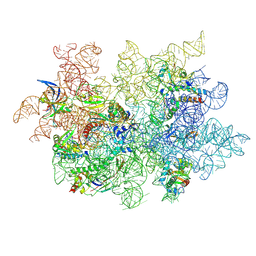 | | WT 12C IM fraction, B-b3 with RluB bound | | Descriptor: | 23S rRNA, Large ribosomal subunit protein bL20, Large ribosomal subunit protein bL21, ... | | Authors: | Lee, J, Sheng, K, Williamson, J.R. | | Deposit date: | 2024-07-10 | | Release date: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.04 Å) | | Cite: | 50S ribosome assembly intermediates at low temperature reveal bound RluB
To Be Published
|
|
9CMZ
 
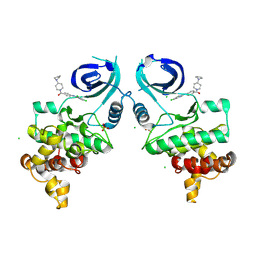 | |
9CPG
 
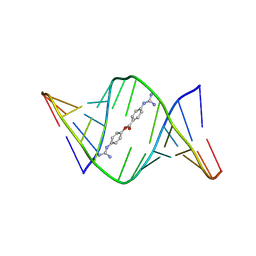 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-carbamimidamidophenyl 4-carbamimidamidobenzoate, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Method: | SOLUTION NMR | | Cite: | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1.
Bioorg.Med.Chem.Lett., 2024
|
|
9CPI
 
 | | Structures of small molecules bound to RNA repeat expansions that cause Huntington's disease-like 2 and myotonic dystrophy type 1 | | Descriptor: | 4-[1-(2-aminoethyl)-4,5-dihydro-1H-imidazol-2-yl]-N-[4-(4,5-dihydro-1H-imidazol-2-yl)phenyl]benzamide, RNA (5'-R(*GP*AP*CP*AP*GP*CP*UP*GP*CP*UP*GP*UP*C)-3') | | Authors: | Chen, J.L, Taghavi, A, Disney, M.D, Fountain, M.A, Childs-Disney, J.L. | | Deposit date: | 2024-07-18 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | SOLUTION NMR | | Cite: | NMR structures of small molecules bound to a model of a CUG RNA repeat expansion.
Bioorg.Med.Chem.Lett., 111, 2024
|
|
9CSG
 
 | | Human Serum Albumin Bound to Cerastecin Compound 5e | | Descriptor: | 2-(4-butylbenzene-1-sulfonamido)-5-(4-{3-carboxy-4-[4-(2-methoxyethyl)benzene-1-sulfonamido]anilino}-4-oxobutanamido)benzoic acid, Albumin, MYRISTIC ACID | | Authors: | Hruza, A, Klein, D.J, Ishchenko, A. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 2024
|
|
9CSI
 
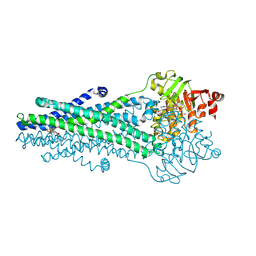 | | A. baumannii MsbA Bound to Cerastecin Compound 5 | | Descriptor: | 3,3'-[(1,4-dioxobutane-1,4-diyl)bis(azanediyl)]bis[(4-butylbenzene-1-sulfonamido)benzoic acid], Lipid A export ATP-binding/permease protein MsbA, MAGNESIUM ION, ... | | Authors: | Klein, D.J, Ishchenko, A, Soisson, S, Cheng, R, Hennig, M. | | Deposit date: | 2024-07-23 | | Release date: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cerastecin Inhibition of the Lipooligosaccharide Transporter MsbA to Combat Acinetobacter baumannii : From Screening Impurity to In Vivo Efficacy.
J.Med.Chem., 2024
|
|
9CTH
 
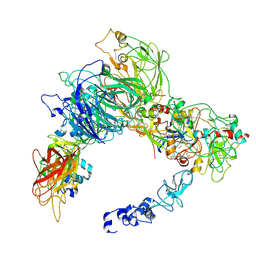 | | Preliminary map of the Prothrombin-prothrombinase complex on nano discs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Activated Factor V (FVa) heavy chain, Activated Factor V (FVa) light chain, ... | | Authors: | Stojanovski, B.M, Mohammed, B.M, Di Cera, E. | | Deposit date: | 2024-07-25 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (6.47 Å) | | Cite: | The Prothrombin-Prothrombinase Interaction.
Subcell Biochem, 104, 2024
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 2024
|
|
9CV0
 
 | | Bufavirus 1 at pH 7.4 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-27 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9CV9
 
 | | Bufavirus 1 at pH 4.0 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-28 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9CWS
 
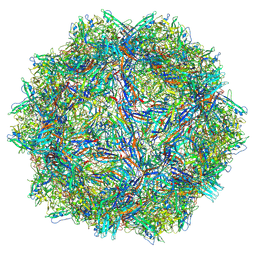 | | Bufavirus 1 at pH 2.6 | | Descriptor: | VP1 | | Authors: | Gulkis, M.C, McKenna, R, Bennett, A.D. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structural Characterization of Human Bufavirus 1: Receptor Binding and Endosomal pH-Induced Changes.
Viruses, 16, 2024
|
|
9D3E
 
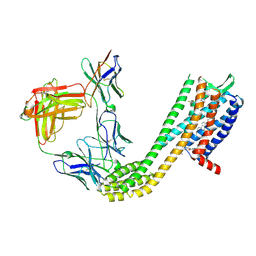 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM2 | | Descriptor: | 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CHOLESTEROL, Human CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-09 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9D3G
 
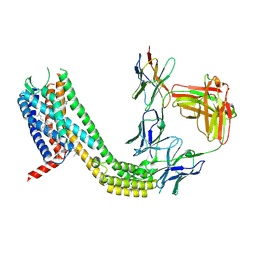 | | Cryo-EM structure of CCR6 bound by SQA1 and OXM1 | | Descriptor: | 1-(4-chlorophenyl)-N-{[(2R)-4-(2,3-dihydro-1H-inden-2-yl)-5-oxomorpholin-2-yl]methyl}cyclopropane-1-carboxamide, 4-[[3,4-bis(oxidanylidene)-2-[[(1~{R})-1-(4-propan-2-ylfuran-2-yl)propyl]amino]cyclobuten-1-yl]amino]-~{N},~{N}-dimethyl-3-oxidanyl-pyridine-2-carboxamide, CCR6, ... | | Authors: | Wasilko, D.J, Wu, H. | | Deposit date: | 2024-08-10 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structural basis for CCR6 modulation by allosteric antagonists.
Nat Commun, 15, 2024
|
|
9D5D
 
 | |






