6Z6N
 
 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6S
 
 | | Crystal structure of Uba4-Urm1 from Chaetomium thermophilum | | Descriptor: | Adenylyltransferase and sulfurtransferase uba4, Ubiquitin-related modifier 1, ZINC ION | | Authors: | Grudnik, P, Pabis, M, Ethiraju Ravichandran, K, Glatt, S. | | Deposit date: | 2020-05-29 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.153 Å) | | Cite: | Molecular basis for the bifunctional Uba4-Urm1 sulfur-relay system in tRNA thiolation and ubiquitin-like conjugation.
Embo J., 39, 2020
|
|
6Z7F
 
 | |
6Z7G
 
 | |
6Z7L
 
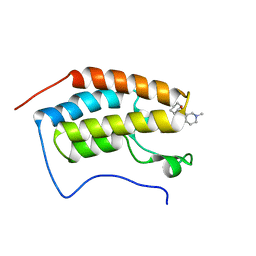 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 with GSK789 | | Descriptor: | (3~{R},4~{R})-~{N}-cyclohexyl-4-[[5-(furan-2-yl)-3-methyl-2-oxidanylidene-1~{H}-1,7-naphthyridin-8-yl]amino]-1-methyl-piperidine-3-carboxamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2020-05-31 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.624 Å) | | Cite: | GSK789: A Selective Inhibitor of the First Bromodomains (BD1) of the Bromo and Extra Terminal Domain (BET) Proteins.
J.Med.Chem., 63, 2020
|
|
6Z7M
 
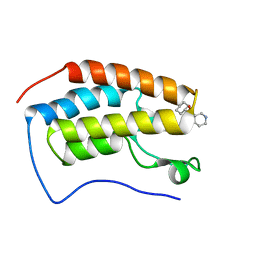 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 (3R,4R)-N-cyclohexyl-4-((3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)amino)piperidine-3-carboxamide | | Descriptor: | (3R,4R)-N-cyclohexyl-4-((3-methyl-2-oxo-1,2-dihydro-1,7-naphthyridin-8-yl)amino)piperidine-3-carboxamide, Bromodomain-containing protein 4 | | Authors: | Chung, C. | | Deposit date: | 2020-05-31 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | GSK789: A Selective Inhibitor of the First Bromodomains (BD1) of the Bromo and Extra Terminal Domain (BET) Proteins.
J.Med.Chem., 63, 2020
|
|
6Z83
 
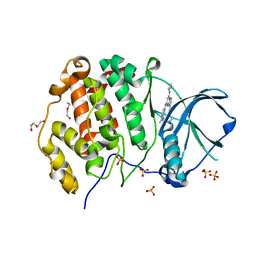 | | CK2 alpha bound to chemical probe SGC-CK2-1 | | Descriptor: | Casein kinase II subunit alpha, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.171 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
6Z84
 
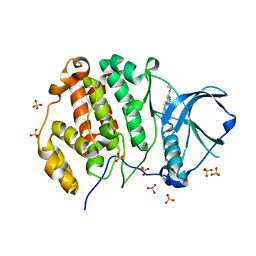 | | CK2 alpha bound to chemical probe SGC-CK2-1 derivative | | Descriptor: | Casein kinase II subunit alpha, SULFATE ION, ~{N}-[1-[3-cyano-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidin-5-yl]indol-6-yl]ethanamide | | Authors: | Kraemer, A, Wells, C, Drewry, D.H, Pickett, J.E, Axtman, A.D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-02 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a potent and selective chemical probe for the pleiotropic kinase CK2.
Cell Chem Biol, 28, 2021
|
|
6Z8N
 
 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule full agonist SRI-9829 | | Descriptor: | (2~{S})-4-methyl-2-[(1-quinolin-8-ylsulfonylindol-3-yl)carbonylamino]pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6Z8P
 
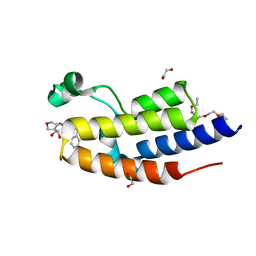 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK973 | | Descriptor: | (2~{S},3~{S})-2-(fluoranylmethyl)-~{N}7-methyl-~{N}5-[(1~{R},5~{S})-3-oxabicyclo[3.1.0]hexan-6-yl]-3-phenyl-2,3-dihydro-1-benzofuran-5,7-dicarboxamide, 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-02 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | GSK973 Is an Inhibitor of the Second Bromodomains (BD2s) of the Bromodomain and Extra-Terminal (BET) Family.
Acs Med.Chem.Lett., 11, 2020
|
|
6ZA8
 
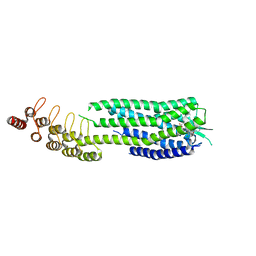 | | Crystal structure of the neurotensin receptor 1 in complex with the small-molecule partial agonist RTI-3a | | Descriptor: | (2~{S})-2-[[1-(7-chloranylquinolin-4-yl)-5-(2,6-dimethoxyphenyl)pyrazol-3-yl]carbonylamino]-4-methyl-pentanoic acid, Neurotensin receptor type 1,Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1),Neurotensin receptor type 1,Neurotensin receptor 1 (NTSR1),Neurotensin receptor 1 (NTSR1) | | Authors: | Deluigi, M, Klipp, A, Hilge, M, Merklinger, L, Klenk, C, Plueckthun, A. | | Deposit date: | 2020-06-05 | | Release date: | 2021-02-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Complexes of the neurotensin receptor 1 with small-molecule ligands reveal structural determinants of full, partial, and inverse agonism.
Sci Adv, 7, 2021
|
|
6ZB0
 
 | |
6ZB1
 
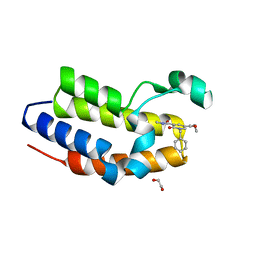 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, ~{N}5-cyclopropyl-~{N}3-methyl-2-oxidanylidene-1-(phenylmethyl)pyridine-3,5-dicarboxamide | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZB2
 
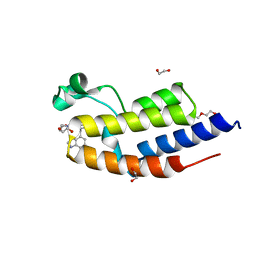 | | C-TERMINAL BROMODOMAIN OF HUMAN BRD2 WITH GSK549 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZB3
 
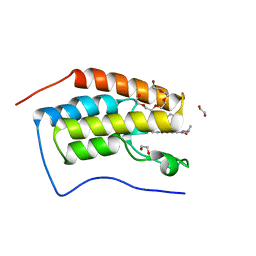 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH GSK620 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, PENTAETHYLENE GLYCOL, ... | | Authors: | Chung, C. | | Deposit date: | 2020-06-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.419 Å) | | Cite: | The Optimization of a Novel, Weak Bromo and Extra Terminal Domain (BET) Bromodomain Fragment Ligand to a Potent and Selective Second Bromodomain (BD2) Inhibitor.
J.Med.Chem., 63, 2020
|
|
6ZBA
 
 | | Crystal structure of PDE4D2 in complex with inhibitor LEO39652 | | Descriptor: | 1,2-ETHANEDIOL, 2-methylpropyl 1-[8-methoxy-5-(1-oxidanylidene-3~{H}-2-benzofuran-5-yl)-[1,2,4]triazolo[1,5-a]pyridin-2-yl]cyclopropane-1-carboxylate, DIMETHYL SULFOXIDE, ... | | Authors: | Akutsu, M, Hakansson, M, Welin, M, Svensson, A, Logan, D.T, Sorensen, M.D. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Early Clinical Development of Isobutyl 1-[8-Methoxy-5-(1-oxo-3 H -isobenzofuran-5-yl)-[1,2,4]triazolo[1,5- a ]pyridin-2-yl]cyclopropanecarboxylate (LEO 39652), a Novel "Dual-Soft" PDE4 Inhibitor for Topical Treatment of Atopic Dermatitis.
J.Med.Chem., 63, 2020
|
|
6ZBP
 
 | | H11-H4 complex with SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H11-H4, SULFATE ION, ... | | Authors: | Naismith, J.H, Huo, J, Mikolajek, H, Ward, P, Dumoux, M, Owens, R.J, LeBas, A. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | H11-D4 complex with SARS-CoV-2 RBD
To Be Published
|
|
6ZDK
 
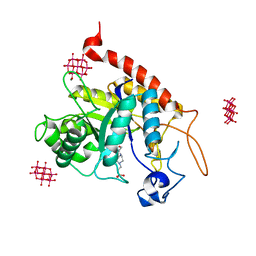 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-tungstotellurate(VI), Glycoprotein endo-alpha-1,2-mannosidase, ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDL
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDM
 
 | | Crystal structure of human heparanase in complex with a N',6O'-bis-sulfated 4-methylumbelliferyl heparan sulfate disaccharide | | Descriptor: | (2~{S},3~{S},4~{R},5~{R},6~{S})-3-[(2~{R},3~{R},4~{R},5~{S},6~{R})-4,5-bis(oxidanyl)-3-(sulfoamino)-6-(sulfooxymethyl)oxan-2-yl]oxy-6-(4-methyl-2-oxidanylidene-chromen-7-yl)oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, L, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-10-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.714 Å) | | Cite: | Structural insights into heparanase activity using a fluorogenic heparan sulfate disaccharide.
Chem.Commun.(Camb.), 56, 2020
|
|
6ZDW
 
 | |
6ZED
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1 | | Descriptor: | 1,2-ETHANEDIOL, 4,5-dimethyl-1~{H}-pyrazolo[3,4-c]pyridazin-3-amine, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F1
To Be Published
|
|
6ZEL
 
 | | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5 | | Descriptor: | 1,2-ETHANEDIOL, 3,5-dimethyl-4-[(6-methylpyrimidin-4-yl)sulfanylmethyl]-1,2-oxazole, Bromodomain-containing protein 4 | | Authors: | Krojer, T, Martinez-Cartro, M, Picaud, S, Filippakopoulos, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Barril, X, von Delft, F. | | Deposit date: | 2020-06-16 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | CRYSTAL STRUCTURE OF THE FIRST BROMODOMAIN OF HUMAN BRD4 IN COMPLEX with compound F5
To Be Published
|
|
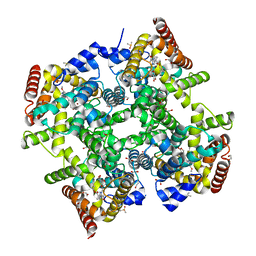
6ZET
 
 | | Crystal structure of proteinase K nanocrystals by electron diffraction with a 20 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.701 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|
6ZEU
 
 | | Crystal structure of proteinase K lamella by electron diffraction with a 50 micrometre C2 condenser aperture | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Evans, G, Zhang, P, Beale, E.V, Waterman, D.G. | | Deposit date: | 2020-06-16 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.004 Å) | | Cite: | A Workflow for Protein Structure Determination From Thin Crystal Lamella by Micro-Electron Diffraction.
Front Mol Biosci, 7, 2020
|
|







