6XQD
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UUC-mRNA, and deacylated P-site tRNA at 2.80A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with sarecycline, UAA-mRNA, and deacylated P-site tRNA at 3.00A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Batool, Z, Lomakin, I.B, Bunick, C.G, Polikanov, Y.S. | | Deposit date: | 2020-07-09 | | Release date: | 2020-08-05 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Sarecycline interferes with tRNA accommodation and tethers mRNA to the 70S ribosome.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6XR8
 
 | | Distinct conformational states of SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Rawson, S, Volloch, S.R, Chen, B. | | Deposit date: | 2020-07-11 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Distinct conformational states of SARS-CoV-2 spike protein.
Science, 369, 2020
|
|
6XTJ
 
 | | The high resolution structure of the FERM domain of human FERMT2 | | Descriptor: | CITRIC ACID, Fermitin family homolog 2,Fermitin family homolog 2,Fermitin family homolog 2 | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-16 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The high resolution structure of the FERM domain of human FERMT2
To Be Published
|
|
6XU6
 
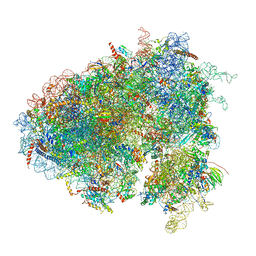 | | Drosophila melanogaster Testis 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU7
 
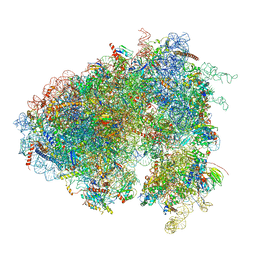 | | Drosophila melanogaster Testis polysome ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XU8
 
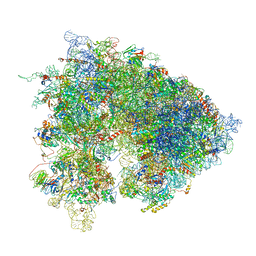 | | Drosophila melanogaster Ovary 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Hopes, T, Agapiou, M, Norris, K, McCarthy, C.G.P, OConnell, M.J, Fontana, J, Aspden, J.L. | | Deposit date: | 2020-01-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome heterogeneity in Drosophila melanogaster gonads through paralog-switching.
Nucleic Acids Res., 50, 2022
|
|
6XV5
 
 | |
6XVX
 
 | |
6XW0
 
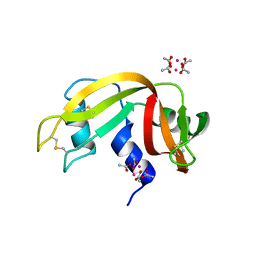 | |
6XWK
 
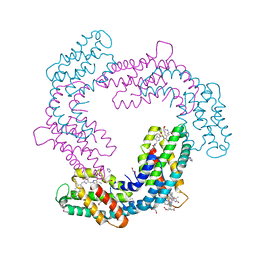 | | Crystal structure of Phormidium rubidum phycocyanin | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Sonani, R.R, Roszak, A.W, Cogdell, R.J, Madamwar, D, Liu, H, Gross, M.L, Blankenship, R.E. | | Deposit date: | 2020-01-23 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Revisiting high-resolution crystal structure of Phormidium rubidum phycocyanin.
Photosyn. Res., 144, 2020
|
|
6XWZ
 
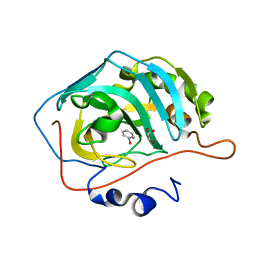 | |
6XXF
 
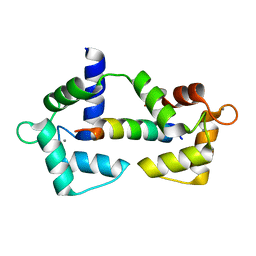 | |
6XXG
 
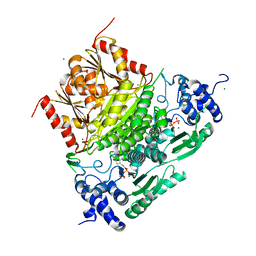 | | Structure of truncated 1-deoxy-D-xylulose 5-phosphate synthase (DXS) from Deinococcus radiodurans | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase,1-deoxy-D-xylulose-5-phosphate synthase, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Gierse, R.M, Reddem, E, Grooves, M.R. | | Deposit date: | 2020-01-27 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Identification of a 1-deoxy-D-xylulose-5-phosphate synthase (DXS) mutant with improved crystallographic properties.
Biochem.Biophys.Res.Commun., 539, 2021
|
|
6XXX
 
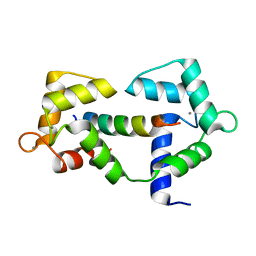 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
6XY3
 
 | |
6XY7
 
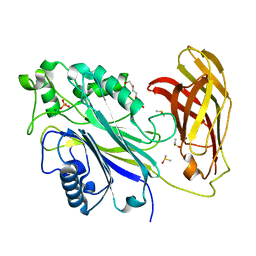 | | Human SHIP1 with magnesium and phosphate bound to the active site | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bradshaw, W.J, Scacioc, A, Fernandez-Cid, A, Mckinley, G, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-01-29 | | Release date: | 2020-02-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Regulation of inositol 5-phosphatase activity by the C2 domain of SHIP1 and SHIP2.
Structure, 2024
|
|
6XYJ
 
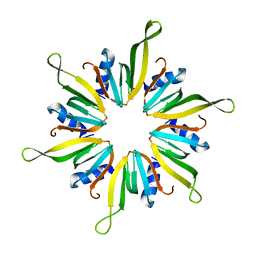 | |
6XYW
 
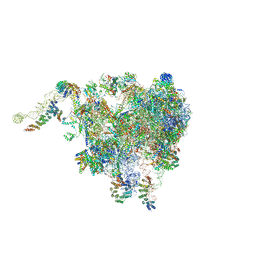 | | Structure of the plant mitochondrial ribosome | | Descriptor: | 28S ribosomal S34 protein, 3-hydroxyisobutyryl-CoA hydrolase-like protein 2, mitochondrial, ... | | Authors: | Soufari, H, Waltz, F, Bochler, A, Giege, P, Hashem, Y. | | Deposit date: | 2020-01-31 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM structure of the RNA-rich plant mitochondrial ribosome.
Nat.Plants, 6, 2020
|
|
6XZA
 
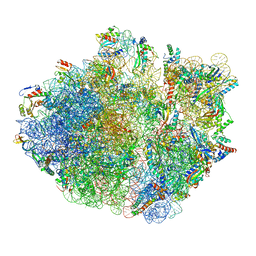 | | E. coli 70S ribosome in complex with dirithromycin, and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6XZB
 
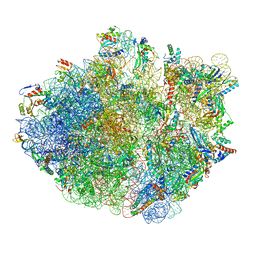 | | E. coli 70S ribosome in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6XZD
 
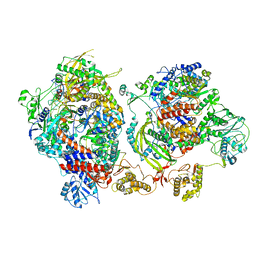 | | Influenza C virus polymerase complex without chicken ANP32A - Subclass 2 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*UP*AP*GP*AP*AP*AP*CP*AP*AP*GP*GP*GP*CP*CP*CP*UP*GP*C)-3'), ... | | Authors: | Keown, J.R, Carrique, L, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-04 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZG
 
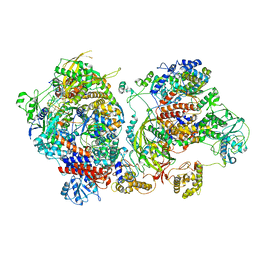 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 3 | | Descriptor: | Influenza viral RNA (vRNA) promoter 47mer, LRRcap domain-containing protein, Polymerase acidic protein, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-04 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZP
 
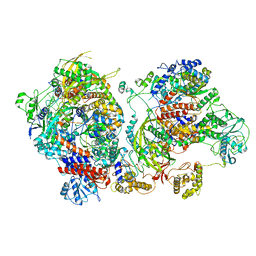 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 4 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6XZR
 
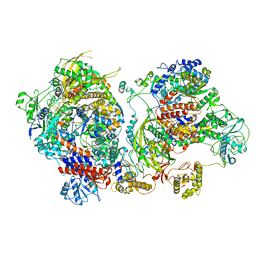 | | Influenza C virus polymerase in complex with chicken ANP32A - Subclass 1 | | Descriptor: | LRRcap domain-containing protein, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Keown, J.R, Fan, H, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|







