8UXF
 
 | |
8UXG
 
 | |
8UXH
 
 | |
8UXI
 
 | |
8UXL
 
 | |
8UXM
 
 | |
8UZT
 
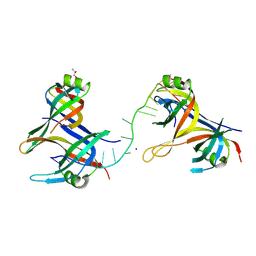 | | Mitochondrial single-stranded binding protein bound to DNA | | Descriptor: | GLYCEROL, SODIUM ION, Single-stranded DNA-binding protein, ... | | Authors: | Riccio, A.A, Pedersen, L.C, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-16 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of the mitochondrial single-stranded DNA binding protein with DNA and DNA polymerase gamma.
Nucleic Acids Res., 2024
|
|
8V3D
 
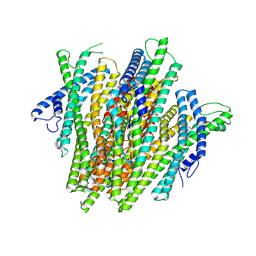 | | AgamOR28 structure bound to 2,4,5-trimethylthiazole | | Descriptor: | 2,4,5-trimethyl-1,3-thiazole, OR28, Odorant receptor Orco | | Authors: | Zhao, J, del Marmol, J. | | Deposit date: | 2023-11-27 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of odor sensing by insect heteromeric odorant receptors.
Science, 384, 2024
|
|
8V3L
 
 | |
8V3N
 
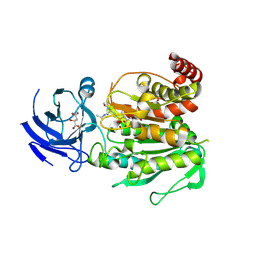 | | CCP5 in complex with Glu-P-Glu transition state analog | | Descriptor: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | Authors: | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | Deposit date: | 2023-11-28 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3T
 
 | |
8V3W
 
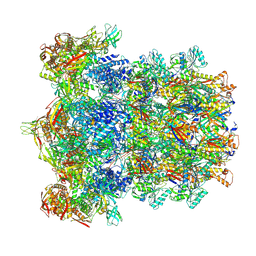 | | CryoEM Structure of Diffocin - precontracted - Baseplate - focused refinement on triplex region | | Descriptor: | Hub-Hydrolase (CD1368), Sheath (CD1363), Sheath initiator (CD1370), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V3Z
 
 | |
8V40
 
 | |
8V41
 
 | | CryoEM Structure of Diffocin - postcontracted - Baseplate - transitional state | | Descriptor: | Sheath (CD1363), Sheath initiator (CD1370), TRI-1 (CD1372), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V43
 
 | | CryoEM Structure of Diffocin - postcontracted - Baseplate - final state | | Descriptor: | Sheath (CD1363), Sheath initiator (CD1370), TRI-1 (CD1372), ... | | Authors: | Cai, X.Y, He, Y, Zhou, Z.H. | | Deposit date: | 2023-11-28 | | Release date: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Atomic structures of a bacteriocin targeting Gram-positive bacteria.
Nat Commun, 15, 2024
|
|
8V4X
 
 | | Structure of MALT1 in complex with an allosteric inhibitor | | Descriptor: | Inhibitor peptide, Mucosa-associated lymphoid tissue lymphoma translocation protein 1, N-{7-[(1S)-1-methoxyethyl]-2-methyl[1,3]thiazolo[5,4-b]pyridin-6-yl}-N'-[6-(2H-1,2,3-triazol-2-yl)-5-(trifluoromethyl)pyridin-3-yl]urea | | Authors: | Judge, R.A, Pappano, W.N. | | Deposit date: | 2023-11-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Inhibition of MALT1 and BCL2 Induces Synergistic Antitumor Activity in Models of B-Cell Lymphoma.
Mol.Cancer Ther., 23, 2024
|
|
8V5A
 
 | |
8V6P
 
 | | Proteus vulgaris tryptophan indole-lyase complexed with 7-aza-L-tryptophan | | Descriptor: | (2E)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)propanoic acid, (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)-L-alanine, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2023-12-02 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Proteus vulgaris tryptophan indole-lyase complexed with L-alanine
To Be Published
|
|
8V7X
 
 | |
8V83
 
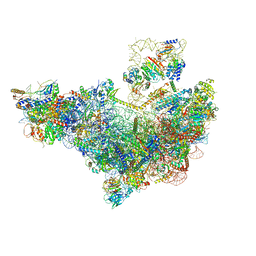 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V8Z
 
 | |
8V9B
 
 | | Lipoprotein(a) Kringle IV domain 7 - Lp(a) KIV7 in complex with LY3441732 | | Descriptor: | (2S,2'S)-3,3'-[carbonylbis(azanediyl-3,1-phenylene)]bis{2-[(3R)-pyrrolidin-1-ium-3-yl]propanoate}, Apolipoprotein(a), MAGNESIUM ION, ... | | Authors: | Hendle, J, Weichert, K, Sauder, J.M. | | Deposit date: | 2023-12-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Discovery of potent small-molecule inhibitors of lipoprotein(a) formation.
Nature, 629, 2024
|
|






