7NDU
 
 | | Gag:02 TCR in complex with HLA-E featuring a non-natural amino acid | | Descriptor: | Beta-2-microglobulin, Gag6V(276-284 H4C), HLA class I histocompatibility antigen, ... | | Authors: | Pengelly, R.J, Robinson, R.A. | | Deposit date: | 2021-02-02 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure-guided stabilization of pathogen-derived peptide-HLA-E complexes using non-natural amino acids conserves native TCR recognition.
Eur.J.Immunol., 52, 2022
|
|
7NE9
 
 | |
7NEO
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 15 | | Descriptor: | 2-cyclobutyl-7-(5-fluoropyridin-3-yl)-5,7-diazaspiro[3.4]octane-6,8-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-02-04 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
7NFX
 
 | | Mammalian ribosome nascent chain complex with SRP and SRP receptor in early state A | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Lee, J.H, Shan, S, Ban, N. | | Deposit date: | 2021-02-08 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Receptor compaction and GTPase rearrangement drive SRP-mediated cotranslational protein translocation into the ER.
Sci Adv, 7, 2021
|
|
7NG8
 
 | |
7NH3
 
 | | Nematocida Huwe1 in open conformation. | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Petrova, O, Grishkovskaya, I, Grabarczyk, D.B, Kessler, D, Haselbach, D, Clausen, T. | | Deposit date: | 2021-02-09 | | Release date: | 2022-03-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.37 Å) | | Cite: | Crystal structure of HUWE1: One ring to ubiquitinate them all
To Be Published
|
|
7NH6
 
 | | Crystal structure of human carbonic anhydrase II with 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide | | Descriptor: | 3-(3-((1-(2-(hydroxymethyl)-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)tetrahydrofuran-3-yl)-1H-1,2,3-triazol-4-yl)methyl)ureido)benzenesulfonamide, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Mechanisms of the Antiproliferative and Antitumor Activity of Novel Telomerase-Carbonic Anhydrase Dual-Hybrid Inhibitors.
J.Med.Chem., 64, 2021
|
|
7NH8
 
 | | Crystal structure of human carbonic anhydrase II with N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide | | Descriptor: | Carbonic anhydrase 2, N-((1-(6-((3aR,7R,7aS)-7-hydroxy-2,2-dimethyltetrahydro-[1,3]dioxolo[4,5-c]pyridin-5(4H)-yl)hexyl)-1H-1,2,3-triazol-4-yl)methyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2021-02-10 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.369 Å) | | Cite: | Synthesis of Azasugar-Sulfonamide conjugates and their Evaluation as Inhibitors of Carbonic Anhydrases: the Azasugar Approach to Selectivity
Eur.J.Org.Chem., 2021
|
|
7NHK
 
 | | LsaA, an antibiotic resistance ABCF, in complex with 70S ribosome from Enterococcus faecalis | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Crowe-McAuliffe, C, Kasari, M, Hauryliuk, V.H, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NHL
 
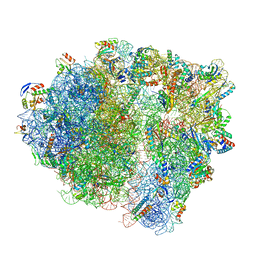 | | VgaA-LC, an antibiotic resistance ABCF, in complex with 70S ribosome from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Murina, V, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NHM
 
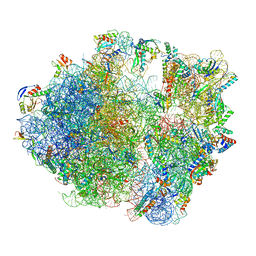 | | 70S ribosome from Staphylococcus aureus | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Murina, V, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NHN
 
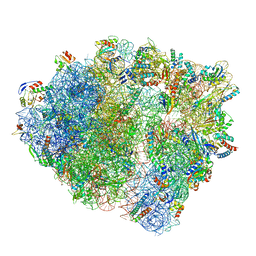 | | VgaL, an antibiotic resistance ABCF, in complex with 70S ribosome from Listeria monocytogenes | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Turnbull, K.J, Hauryliuk, V, Wilson, D.N. | | Deposit date: | 2021-02-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens.
Nat Commun, 12, 2021
|
|
7NKT
 
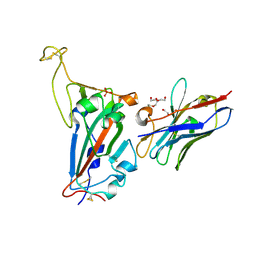 | | RBD domain of SARS-CoV2 in complex with neutralizing nanobody NM1226 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Ostertag, E, Zocher, G, Stehle, T. | | Deposit date: | 2021-02-18 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NeutrobodyPlex-monitoring SARS-CoV-2 neutralizing immune responses using nanobodies.
Embo Rep., 22, 2021
|
|
7NLV
 
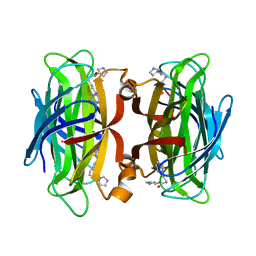 | | WILDTYPE CORE-STREPTAVIDIN WITH a conjugated BIOTINYLATED PYRROLIDINE II | | Descriptor: | 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl)-N-((S)-pyrrolidin-3-yl)pentanamide, Streptavidin | | Authors: | Nodling, A.R, Santi, N, Tsai, Y.H, Rizkallah, P, Luk, L.Y.P, Jin, Y. | | Deposit date: | 2021-02-22 | | Release date: | 2022-03-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The role of streptavidin and its variants in catalysis by biotinylated secondary amines.
Org.Biomol.Chem., 19, 2021
|
|
7NM7
 
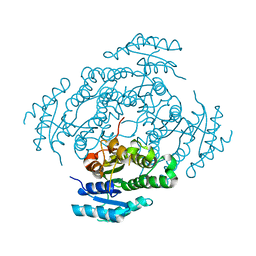 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM | | Descriptor: | Antimycin pathway standalone ketoreductase enzyme, AntM | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
7NM8
 
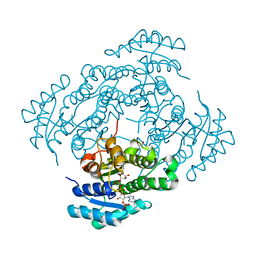 | | The crystal structure of the antimycin pathway standalone ketoreductase, AntM, in complex with NADPH | | Descriptor: | Antimycin pathway standalone ketoreductase, AntM, GLYCEROL, ... | | Authors: | Fazal, A, Hemsworth, G.R, Webb, M.E, Seipke, R.F. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A Standalone beta-Ketoreductase Acts Concomitantly with Biosynthesis of the Antimycin Scaffold.
Acs Chem.Biol., 16, 2021
|
|
7NMC
 
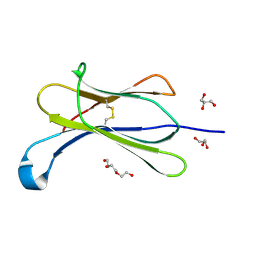 | | Crystal structure of beta-2-microglobulin D76E mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL, TRIETHYLENE GLYCOL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMO
 
 | | Crystal structure of beta-2-microglobulin D76A mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMR
 
 | | Crystal structure of beta-2-microglobulin D76S mutant | | Descriptor: | Beta-2-microglobulin, GLYCEROL | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMT
 
 | | Crystal structure of beta-2-microglobulin D76G mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMU
 
 | |
7NMV
 
 | | Crystal structure of beta-2-microglobulin D76Q mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMY
 
 | | Crystal structure of beta-2-microglobulin D76Y mutant | | Descriptor: | Beta-2-microglobulin | | Authors: | Guthertz, N. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The effect of mutation on an aggregation-prone protein: An in vivo, in vitro, and in silico analysis.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7NMZ
 
 | |







