7FCP
 
 | |
7FCQ
 
 | |
7WHC
 
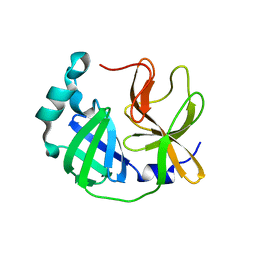 | | Crystal structure of SARS-CoV-2 3CLpro catalytic domain | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Shin, D.H, Jo, S.R. | | Deposit date: | 2021-12-30 | | Release date: | 2022-06-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.269 Å) | | Cite: | Dimerization Tendency of 3CLpros of Human Coronaviruses Based on the X-ray Crystal Structure of the Catalytic Domain of SARS-CoV-2 3CLpro.
Int J Mol Sci, 23, 2022
|
|
7XIC
 
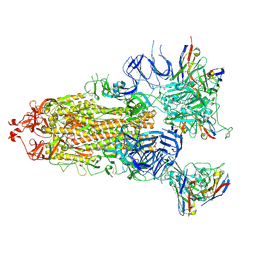 | | S-ECD (Omicron) in complex with STS165 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XID
 
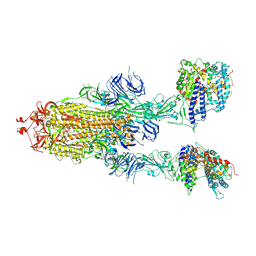 | | S-ECD (Omicron) in complex with PD of ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, Y.N, Shen, Y.P, Zhang, Y.Y, Yan, R.H. | | Deposit date: | 2022-04-12 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and functional analysis of an inter-Spike bivalent neutralizing antibody against SARS-CoV-2 variants.
Iscience, 25, 2022
|
|
7XO5
 
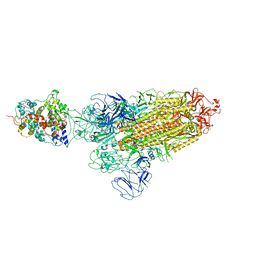 | | SARS-CoV-2 Omicron BA.1 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO6
 
 | | SARS-CoV-2 Omicron BA.1 Variant RBD with mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XO9
 
 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOA
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with one mouse ACE2 Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOC
 
 | | SARS-CoV-2 Omicron BA.2 Variant RBD complexed with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7XOD
 
 | | SARS-CoV-2 Omicron BA.2 Variant Spike Trimer with three JMB2002 Fab Bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of JMB2002 Fab, ... | | Authors: | Xu, Y, Wu, C, Liu, H, Yin, W, Xu, H.E. | | Deposit date: | 2022-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins.
Cell Res., 32, 2022
|
|
7F3Q
 
 | | SARS-CoV-2 RBD in complex with A5-10 Fab and A34-2 Fab | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of A34-2 Fab, ... | | Authors: | Dou, Y, Wang, X, Liu, P, Lu, B, Wang, K. | | Deposit date: | 2021-06-16 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Development of neutralizing antibodies against SARS-CoV-2, using a high-throughput single-B-cell cloning method.
Antib Ther, 6, 2023
|
|
7F5R
 
 | | Crystal structure of SARS-CoV-2 Y453F-RBD bound to mink ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Wang, X.Q, Ding, Q, Lan, J, Ren, W.L. | | Deposit date: | 2021-06-22 | | Release date: | 2022-06-22 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Mutation Y453F in the spike protein of SARS-CoV-2 enhances interaction with the mink ACE2 receptor for host adaption.
Plos Pathog., 17, 2021
|
|
7FC5
 
 | | Crystal structure of SARS-CoV-2 RBD and horse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike protein S1 | | Authors: | Wang, X.Q, Lan, J, Ge, J.W. | | Deposit date: | 2021-07-13 | | Release date: | 2022-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Structure, 30, 2022
|
|
7MB4
 
 | |
7MB5
 
 | |
7MB6
 
 | |
7MB7
 
 | |
7MB8
 
 | |
7MB9
 
 | |
7T70
 
 | |
7T8M
 
 | |
7T8R
 
 | |
7T9Y
 
 | |
7TA4
 
 | |








