5RVQ
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002508153 | | Descriptor: | 5-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVR
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000016052862 | | Descriptor: | 1H-indazole, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVS
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000000159004 | | Descriptor: | 5-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002582714 | | Descriptor: | 6-methyl-1H-indole-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVU
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000002506130 | | Descriptor: | 6-phenylpyridine-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
5RVV
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000020269197 | | Descriptor: | 6-methyl-1H-indole-3-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.C, Young, I.D, Thompson, M.C, Fraser, J.S. | | Deposit date: | 2020-10-02 | | Release date: | 2020-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7B3O
 
 | | Crystal structure of the SARS-CoV-2 RBD in complex with STE90-C11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of Fab Fragment, Light Chain of Fab Fragment, ... | | Authors: | Kluenemann, T, Van den Heuvel, J. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A SARS-CoV-2 neutralizing antibody selected from COVID-19 patients binds to the ACE2-RBD interface and is tolerant to most known RBD mutations.
Cell Rep, 36, 2021
|
|
7CAB
 
 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CWM
 
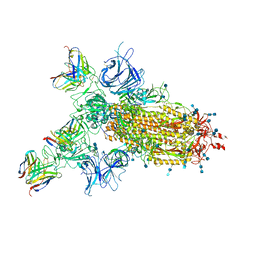 | |
7CWN
 
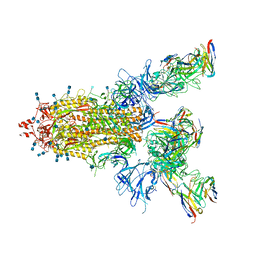 | | P17-H014 Fab cocktail in complex with SARS-CoV-2 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, N, Wang, X. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CWO
 
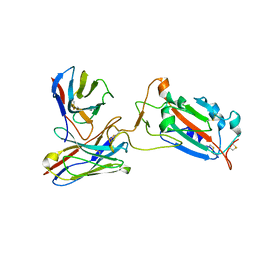 | | SARS-CoV-2 spike protein RBD and P17 fab complex | | Descriptor: | Spike glycoprotein, heavy chain of P17 Fab, light chain of P17 Fab | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-29 | | Release date: | 2020-12-16 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Rational development of a human antibody cocktail that deploys multiple functions to confer Pan-SARS-CoVs protection.
Cell Res., 31, 2021
|
|
7CWS
 
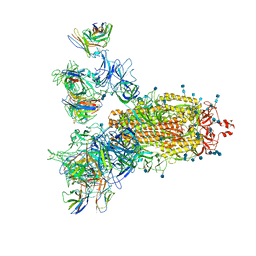 | | SARS-CoV-2 Spike Proteins Trimer in Complex with FC05 and H014 Fabs Cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy Chain of FC05 Fab, ... | | Authors: | Wang, L, Wang, X. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7CWU
 
 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
7DF3
 
 | | SARS-CoV-2 S trimer, S-closed | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DF4
 
 | | SARS-CoV-2 S-ACE2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Cong, X, Yao, C. | | Deposit date: | 2020-11-06 | | Release date: | 2020-12-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7DK3
 
 | | SARS-CoV-2 S trimer, S-open | | Descriptor: | Spike glycoprotein | | Authors: | Xu, C, Cong, Y. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Conformational dynamics of SARS-CoV-2 trimeric spike glycoprotein in complex with receptor ACE2 revealed by cryo-EM.
Sci Adv, 7, 2021
|
|
7KNE
 
 | | Cryo-EM structure of single ACE2-bound SARS-CoV-2 trimer spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNH
 
 | | Cryo-EM Structure of Double ACE2-Bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KNI
 
 | | Cryo-EM structure of Triple ACE2-bound SARS-CoV-2 Trimer Spike at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Rapp, M, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-04 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
7KX5
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with noncovalent inhibitor Jun8-76-3A | | Descriptor: | 3C-like proteinase, GLYCEROL, N-([1,1'-biphenyl]-4-yl)-N-[(1R)-2-oxo-2-{[(1S)-1-phenylethyl]amino}-1-(pyridin-3-yl)ethyl]furan-2-carboxamide | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2020-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity.
J.Am.Chem.Soc., 143, 2021
|
|
7KYU
 
 | | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate | | Descriptor: | 1,2-ETHANEDIOL, 1-[(1H-indole-5-carbonyl)oxy]-1H-benzotriazole, 3C-like proteinase | | Authors: | Tan, K, Maltseva, N.I, Jedrzejczak, R.P, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-12-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | The crystal structure of SARS-CoV-2 Main Protease with the formation of Cys145-1H-indole-5-carboxylate
To Be Published
|
|
6ZLR
 
 | |
7B3B
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -3 (structure 1) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*G)-D(P*(RMP))-R(P*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3C
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with Remdesivir at position -4 (structure 2) | | Descriptor: | DNA/RNA (5'-R(P*CP*UP*AP*CP*GP*CP*A)-D(P*(RMP))-R(P*GP*UP*G)-3'), Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | Authors: | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | Deposit date: | 2020-11-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|








