8ZYF
 
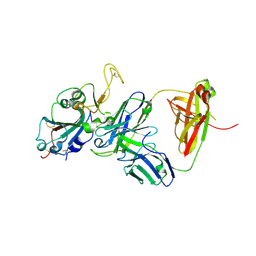 | | Crystal structure of ZW2G10 Fab in complex with omicron RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ZW2G10 fab heavy chain, ... | | Authors: | Yuan, H.Y, Li, J.M. | | Deposit date: | 2024-06-17 | | Release date: | 2025-03-26 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nasal delivery of secretory IgA confers enhanced neutralizing activity against Omicron variants compared to its IgG counterpart.
Mol.Ther., 33, 2025
|
|
9D2K
 
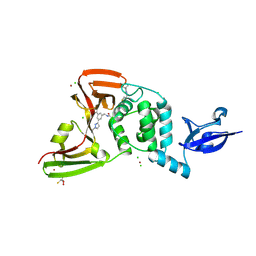 | | SARS-CoV-2 Papain-like Protease (PLpro) complex with covalent inhibitor Jun13567 | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-(3-{2-[4-(3,3-dimethylazetidin-1-yl)-4-oxobutanoyl]hydrazin-1-yl}-3-oxopropyl)-N-{(1R)-1-[(3P,5P)-3-(1-ethyl-1H-pyrazol-3-yl)-5-(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}benzamide, CHLORIDE ION, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2024-08-08 | | Release date: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-Based Design of Covalent SARS-CoV-2 Papain-like Protease Inhibitors.
J.Med.Chem., 67, 2024
|
|
9GXE
 
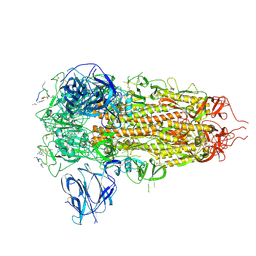 | | Structure of the SARS-CoV spike glycoprotein in complex with a homotrimeric Bicycle molecule | | Descriptor: | 2,4,6-tris(chloromethyl)-1,3,5-triazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Drulyte, I, Pellegrino, S, Harman, M, Bezerra, G.A. | | Deposit date: | 2024-09-30 | | Release date: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structure of the SARS-CoV spike glycoprotein in complex with a homotrimeric Bicycle molecule
To Be Published
|
|
9GXG
 
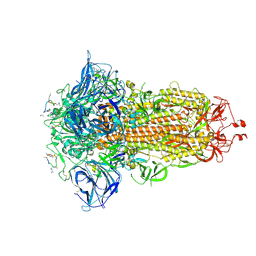 | | Structure of the SARS-CoV spike glycoprotein in complex with a biparatopic Bicycle molecule | | Descriptor: | 1-[3,5-bis(2-chloranylethanoyl)-1,3,5-triazinan-1-yl]-2-chloranyl-ethanone, 2,4,6-tris(chloromethyl)-1,3,5-triazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Drulyte, I, Pellegrino, S, Harman, M, Bezerra, G.A. | | Deposit date: | 2024-09-30 | | Release date: | 2025-03-26 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Structure of the SARS-CoV spike glycoprotein in complex with a biparatopic Bicycle molecule
To Be Published
|
|
8YWE
 
 | |
8ZFV
 
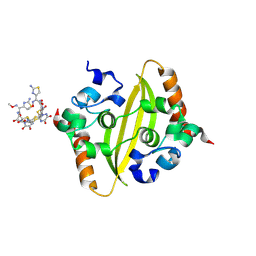 | | Crystal Structure of C-terminal domain of nucleocapsid protein from SARS-CoV-2 in complex with ceftriaxone | | Descriptor: | Ceftriaxone, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Dhaka, P, Mahto, J.K, Tomar, S, Kumar, P. | | Deposit date: | 2024-05-08 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the RNA binding inhibitors of the C-terminal domain of the SARS-CoV-2 nucleocapsid.
J.Struct.Biol., 217, 2025
|
|
9IN1
 
 | |
9KR5
 
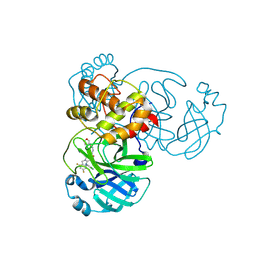 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 3 | | Descriptor: | (6~{E})-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-[(3~{S})-oxolan-3-yl]oxypyridin-3-yl]-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-27 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSH
 
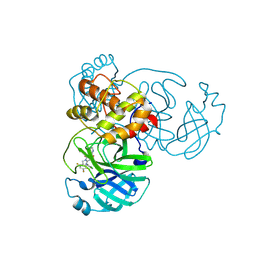 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 1 | | Descriptor: | 3C-like proteinase nsp5, 6-[(6-chloranyl-2-methyl-indazol-5-yl)amino]-3-pyridin-3-yl-1-[[2,4,5-tris(fluoranyl)phenyl]methyl]-1,3,5-triazine-2,4-dione | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSI
 
 | | Crystal Structure of SARS-CoV-2 main protease in complex with compound 5 | | Descriptor: | (6E)-1-[[5-chloranyl-4-fluoranyl-2-(4-fluoranylphenoxy)phenyl]methyl]-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methoxypyridin-3-yl)-1,3,5-triazinane-2,4-dione, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSJ
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 8 | | Descriptor: | 3-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-[5-(2-methoxyethoxy)pyridin-3-yl]-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-4-methyl-benzenecarbonitrile, 3C-like proteinase nsp5 | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9KSK
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with compound 10 | | Descriptor: | 3C-like proteinase, 4-[4-chloranyl-2-[[(6E)-6-(6-chloranyl-2-methyl-indazol-5-yl)imino-3-(5-methylpyridin-3-yl)-2,4-bis(oxidanylidene)-1,3,5-triazinan-1-yl]methyl]-5-fluoranyl-phenoxy]-2-fluoranyl-benzenecarbonitrile | | Authors: | Zhong, Y, Zhao, L, Zhang, W, Peng, W. | | Deposit date: | 2024-11-29 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expanding the utilization of binding pockets proves to be effective for noncovalent small molecule inhibitors against SARS-CoV-2 M pro.
Eur.J.Med.Chem., 289, 2025
|
|
9LBS
 
 | | Cryo-EM structure of Omicron BA.1 RBD complexed with ConBA-998,S309 and S304Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of ConBA-998, Heavy chain of S304 Fab, ... | | Authors: | Zhang, Z, Ju, B, Liu, C. | | Deposit date: | 2025-01-03 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Cryo-EM structure of Omicron BA.1 RBD complexed with ConBA-998,S309 and S304Fabs
To Be Published
|
|
9LDS
 
 | |
9LYO
 
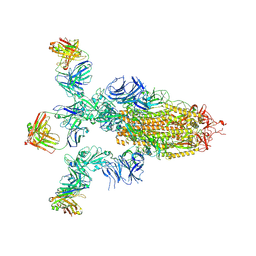 | | Alpha SARS-CoV-2 spike protein in complex with REGN10987 Fab homologue. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, REGN10987 Fab homologue (Heavy chain), ... | | Authors: | Kocharovskaya, M.V, Pichkur, E.B, Shenkarev, Z.O, Lyukmanova, E.N. | | Deposit date: | 2025-02-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structure and dynamics of Alpha B.1.1.7 SARS-CoV-2 S-protein in complex with Fab of neutralizing antibody REGN10987.
Biochem.Biophys.Res.Commun., 755, 2025
|
|
9LYP
 
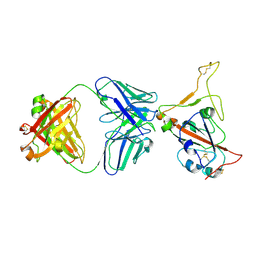 | | Alpha SARS-CoV-2 spike protein RBD-down in complex with REGN10987 Fab homologue (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, REGN10987 Fab homologue (Heavy chain), REGN10987 Fab homologue (Light chain), ... | | Authors: | Kocharovskaya, M.V, Pichkur, E.B, Shenkarev, Z.O, Lyukmanova, E.N. | | Deposit date: | 2025-02-20 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of Alpha B.1.1.7 SARS-CoV-2 S-protein in complex with Fab of neutralizing antibody REGN10987.
Biochem.Biophys.Res.Commun., 755, 2025
|
|
9Q8H
 
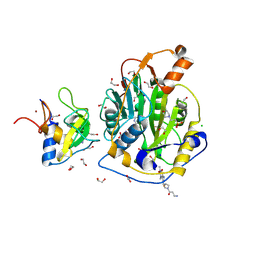 | | SARS-CoV-2 methyltransferase nsp10-16 in complex with SAM and theophylline derivative BDH 34019023 | | Descriptor: | 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Kremling, V, Sprenger, J, Turk, D, Oberthuer, D, Kiene, A. | | Deposit date: | 2025-02-24 | | Release date: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Crystal structures of SARS-CoV-2 methyltransferase nsp10-16 with Cap0-site binders
To Be Published
|
|
8YWY
 
 | | Crystal structure of SARS-Cov-2 main protease E166N mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YWZ
 
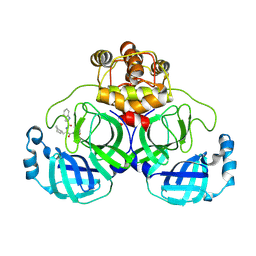 | | Crystal structure of SARS-Cov-2 main protease H163A mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zou, X.F, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-04-01 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9CYB
 
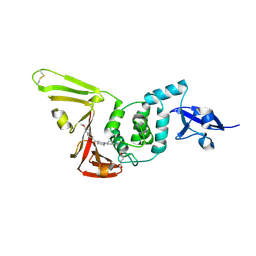 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P1 | | Descriptor: | Papain-like protease, SUCCINIC ACID, [(3R)-1-cyclopentylpiperidin-3-yl](6-methoxynaphthalen-2-yl)methanone | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYC
 
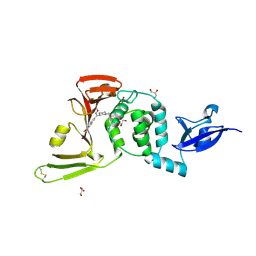 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P2 | | Descriptor: | (E)-1-[(3R)-1-cyclopentylpiperidin-3-yl]-N-methoxy-1-(6-methoxynaphthalen-2-yl)methanimine, ACETIC ACID, GLYCEROL, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchel, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-01 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYD
 
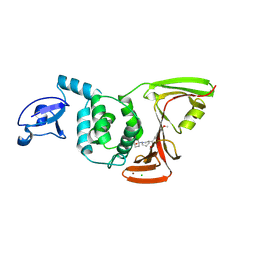 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P4 | | Descriptor: | (1S,4s)-4-{(3R)-3-[(E)-(methoxyimino)(6-methoxynaphthalen-2-yl)methyl]piperidin-1-yl}cyclohexan-1-ol, CHLORIDE ION, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9CYK
 
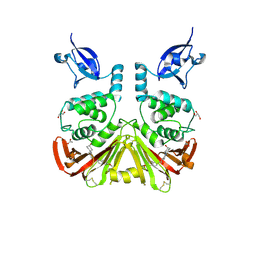 | | SARS-CoV-2 PLpro in complex with inhibitor WEHI-P24 | | Descriptor: | ACETIC ACID, GLYCEROL, Papain-like protease, ... | | Authors: | Calleja, D.J, Lechtenberg, B.C, Kuchel, N.W, Devine, S.M, Bader, S.M, Doerflinger, M, Mitchell, J.P, Lessene, G, Komander, D. | | Deposit date: | 2024-08-02 | | Release date: | 2025-04-09 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A novel PLpro inhibitor improves outcomes in a pre-clinical model of long COVID.
Nat Commun, 16, 2025
|
|
9INL
 
 | | Crystal structure of SARS-Cov-2 main protease E166R mutant in complex with Bofutrelvir | | Descriptor: | Replicase polyprotein 1a, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2024-07-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
9LUF
 
 | | Crystal structure of SARS-Cov-2 main protease S144A mutant in complex with Bofutrelvir | | Descriptor: | ORF1a polyprotein, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zhou, X.L, Li, J. | | Deposit date: | 2025-02-08 | | Release date: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|








