8HC5
 
 | | SARS-CoV-2 wildtype S1 in complex with YB9-258 Fab and R1-32 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of R1-32 Fab, Heavy chain of YB9-258 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC6
 
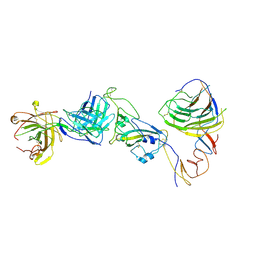 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB9-258 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB9-258 Fab, Light chain of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.69 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC7
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) complex with YB9-258 Fab, focused refinement of RBD-dimer region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain variable region of YB9-258, Light chain variable region of YB9-258, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.54 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC8
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with YB13-292 Fab, focused refinement of Fab region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, Light chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HC9
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (3 RBD down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (6.03 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCA
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (1 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
8HCB
 
 | | SARS-CoV-2 Omicron BA.1 spike trimer (6P) in complex with 3 YB13-292 Fabs (2 RBD up) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of YB13-292 Fab, ... | | Authors: | Liu, B, Gao, X, Chen, Q, Li, Z, Su, M, He, J, Xiong, X. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-25 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Somatically hypermutated antibodies isolated from SARS-CoV-2 Delta infected patients cross-neutralize heterologous variants.
Nat Commun, 14, 2023
|
|
7WRH
 
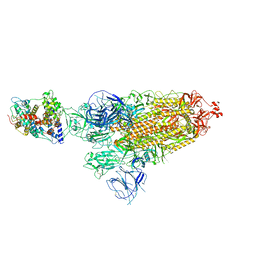 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Han, P, Xie, Y, Qi, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Broader-species receptor binding and structural bases of Omicron SARS-CoV-2 to both mouse and palm-civet ACE2s.
Cell Discov, 8, 2022
|
|
7WZO
 
 | |
7YCK
 
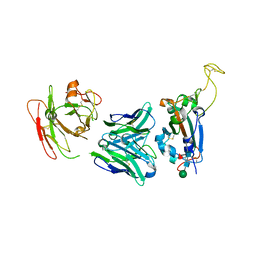 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with FP-12A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, FP-12A Fab light chain, ... | | Authors: | Nguyen, V.H.T, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCL
 
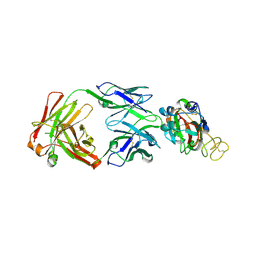 | | Crystal structure of SARS-CoV-2 Spike RBD in complex with IS-9A Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, IS-9A Fab light chain, ... | | Authors: | Mohapatra, A, Chen, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7YCN
 
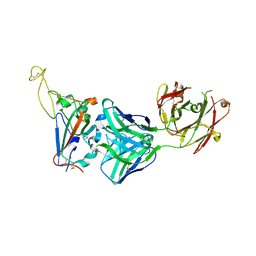 | |
8HHX
 
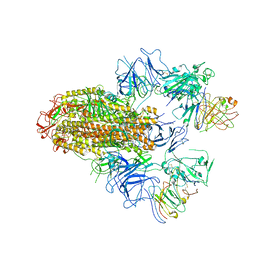 | | SARS-CoV-2 Delta Spike in complex with FP-12A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FP-12A Fab heavy chain, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8HHY
 
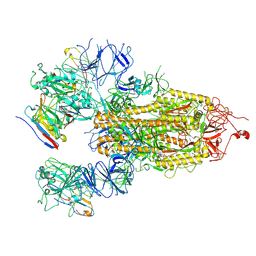 | | SARS-CoV-2 Delta Spike in complex with IS-9A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IS-9A Fab heavy chain, ... | | Authors: | Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
8HHZ
 
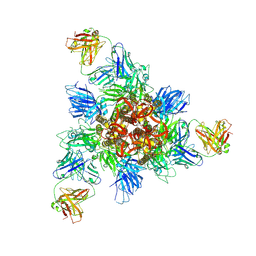 | | SARS-CoV-2 Omicron BA.1 Spike in complex with IY-2A | | Descriptor: | IY-2A Fab heavy chain, IY-2A Fab light chain, Spike glycoprotein | | Authors: | Chen, X, Mohapatra, A, Wu, Y.-M. | | Deposit date: | 2022-11-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structural basis for a conserved neutralization epitope on the receptor-binding domain of SARS-CoV-2.
Nat Commun, 14, 2023
|
|
7TPK
 
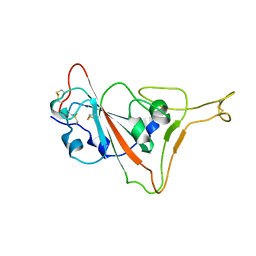 | |
7XWX
 
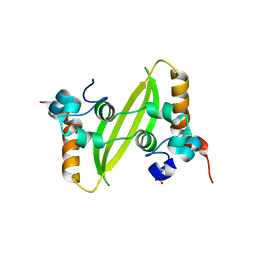 | | Crystal structure of SARS-CoV-2 N-CTD | | Descriptor: | Nucleoprotein, PHOSPHATE ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XWZ
 
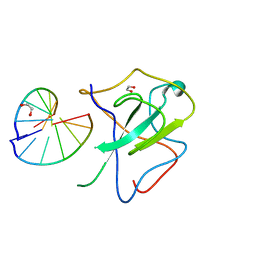 | | Crystal structure of SARS-CoV-2 N-NTD and dsRNA complex | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleoprotein, ... | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7XX1
 
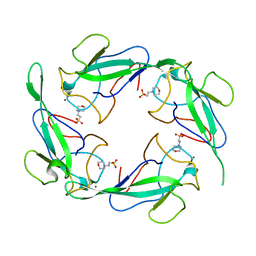 | | Crystal structure of SARS-CoV-2 N-NTD | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Nucleoprotein, ZINC ION | | Authors: | Luan, X.D, Li, X.M, Li, Y.F. | | Deposit date: | 2022-05-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Antiviral drug design based on structural insights into the N-terminal domain and C-terminal domain of the SARS-CoV-2 nucleocapsid protein.
Sci Bull (Beijing), 67, 2022
|
|
7YUE
 
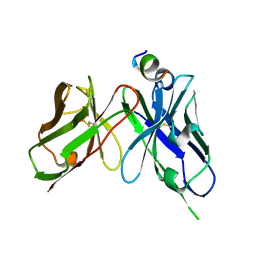 | | Epitope-directed anti-SARS CoV 2 scFv engineered against the key spike protein region. | | Descriptor: | Single chain variable Fragment, Spike protein S2 | | Authors: | Kumar, U, Jaiswal, D, Gaur, V, Salunke, D.M. | | Deposit date: | 2022-08-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Epitope-directed anti-SARS-CoV-2 scFv engineered against the key spike protein region could block membrane fusion.
Protein Sci., 32, 2023
|
|
8DM5
 
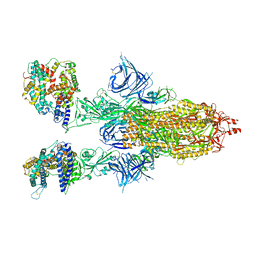 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM6
 
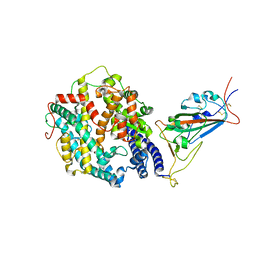 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM7
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DM9
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|








