8XRQ
 
 | | SARS-CoV-2 BA.1 spike RBD in complex bound with VacBB-639 | | Descriptor: | Heavy chain of VacBB 639 Fab, Light chain of VacBB 639 Fab, Spike protein S1 | | Authors: | Liu, C.C, Ju, B, Zhang, Z. | | Deposit date: | 2024-01-08 | | Release date: | 2024-12-18 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Rapid clonal expansion and somatic hypermutation contribute to the fate of SARS-CoV-2 broadly neutralizing antibodies.
J Immunol., 214, 2025
|
|
9BIH
 
 | | SARS-CoV-2 endoribonuclease Nsp15 bound to dsRNA with 1 nucleotide bulge | | Descriptor: | RNA (34-mer), RNA (35-mer), Uridylate-specific endoribonuclease nsp15 | | Authors: | Wright, Z.M, Butay, K.J, Krahn, J.M, Borgnia, M.J, Stanley, R.E. | | Deposit date: | 2024-04-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Spontaneous base flipping helps drive Nsp15's preferences in double stranded RNA substrates.
Nat Commun, 16, 2025
|
|
8ZPP
 
 | | Local CryoEM structure of the SARS-CoV-2 BA.5 in complex with ORB10 Fab | | Descriptor: | Spike glycoprotein,Fibritin, variable heavy chain of ORB10 Fab, variable light chain of ORB10 Fab | | Authors: | Cao, S, Leng, C, Hu, H. | | Deposit date: | 2024-05-30 | | Release date: | 2024-12-25 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into hybridoma-derived neutralizing monoclonal antibodies against Omicron BA.5 and XBB.1.16 variants of SARS-CoV-2.
J.Virol., 99, 2025
|
|
9BF9
 
 | | Human LAG-3-HLA-DR1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, HLA class II histocompatibility antigen DR beta chain, ... | | Authors: | Petersen, J, Rossjohn, J. | | Deposit date: | 2024-04-17 | | Release date: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the human LAG-3-HLA-DR1-peptide complex.
Sci Immunol, 9, 2024
|
|
8RJ5
 
 | | P1-15 T-cell Receptor bound to HLA A*2402-NF9 pMHC complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, P1-15 T-cell Receptor Alpha Chain, ... | | Authors: | Wall, A, Sewell, A.K, Motozono, C, Rizkallah, P.J, Fuller, A. | | Deposit date: | 2023-12-20 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | NF9 T-cell Receptor bound to HLA A*2402-NF9 pMHC complex (Paper Pending)
To Be Published
|
|
8RJH
 
 | | HLA A*2402-NF9_6F pMHC complex | | Descriptor: | ASN-TYR-ASN-TYR-LEU-PHE-ARG-LEU-PHE, Beta-2-microglobulin, MHC class I antigen | | Authors: | Wall, A, Sewell, A.K, Motozono, C, Rizkallah, P.J, Fuller, A. | | Deposit date: | 2023-12-21 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | HLA A*2402-NF9_6F pMHC complex
To Be Published
|
|
8RJI
 
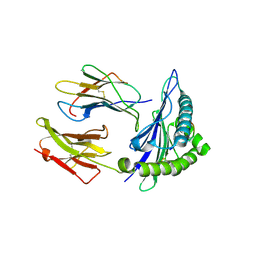 | | HLA A*2402-NF9_5R pMHC complex | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Spike glycoprotein | | Authors: | Wall, A, Motozono, C, Sewell, A.K, Rizkallah, P.J, Fuller, A. | | Deposit date: | 2023-12-21 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HLA A*2402-NF9_5R pMHC complex
To Be Published
|
|
8Y4D
 
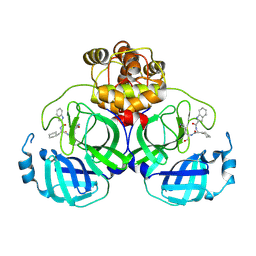 | |
8Y4G
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Zeng, P, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-01-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8Y4H
 
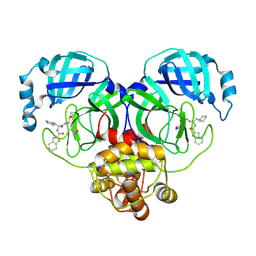 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with Bofutrelvir | | Descriptor: | 3C-like proteinase nsp5, ~{N}-[(2~{S})-3-cyclohexyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]propan-2-yl]-1~{H}-indole-2-carboxamide | | Authors: | Guo, L, Wang, W.W, Zhang, J, Li, J. | | Deposit date: | 2024-01-30 | | Release date: | 2025-01-08 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Inhibitory efficacy and structural insights of Bofutrelvir against SARS-CoV-2 M pro mutants and MERS-CoV M pro.
Commun Biol, 8, 2025
|
|
8YDP
 
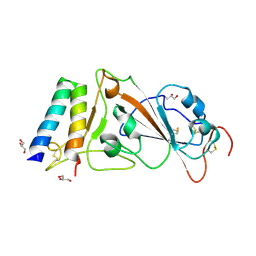 | | Crystal structure of the receptor binding domain of SARS-CoV-2 spike protein in complex with Ce9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SARS-CoV-2 inhibiting peptide Ce9, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDQ
 
 | |
8YDR
 
 | |
8YDS
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant spike protein in complex with Ce59 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SARS-CoV-2 inhibiting peptide Ce59, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDT
 
 | |
8YDU
 
 | |
8YDV
 
 | |
8YDW
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron XBB.1.5 variant spike protein in complex with CeSPIACE | | Descriptor: | GLYCEROL, SARS-CoV-2 inhibiting peptide CeSPIACE, SODIUM ION, ... | | Authors: | Nakamura, S, Numoto, N, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDX
 
 | | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.2 variant) in complex with CeSPIACE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CeSPIACE, Spike glycoprotein | | Authors: | Suzuki, H, Nakamura, S, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8YDZ
 
 | | Cryo-EM structure of SARS-CoV-2 spike ectodomain (HexaPro, Omicron BA.5 variant) in complex with CeSPIACE, class 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CeSPIACE, Spike glycoprotein | | Authors: | Suzuki, H, Nakamura, S, Fujiyoshi, Y. | | Deposit date: | 2024-02-21 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | Structure-guided engineering of a mutation-tolerant inhibitor peptide against variable SARS-CoV-2 spikes.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9IUP
 
 | | KP.3 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L. | | Deposit date: | 2024-07-22 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Cell Discov, 10, 2024
|
|
9IUQ
 
 | | KP.2 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Feng, L.L. | | Deposit date: | 2024-07-22 | | Release date: | 2025-01-15 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Cell Discov, 10, 2024
|
|
9IUU
 
 | | JN.1 RBD with Q493E in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | Authors: | Feng, L.L. | | Deposit date: | 2024-07-22 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Cell Discov, 10, 2024
|
|
9JJ7
 
 | |
8XAL
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein (Fragment), ... | | Authors: | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2023-12-04 | | Release date: | 2025-01-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|








