8XLV
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P), 1-RBD-up state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-26 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XM5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMG
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XMT
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P), RBD-closed state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN2
 
 | | SARS-CoV-2 Omicron EG.5.1 RBD in complex with human ACE2 (local refined from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN3
 
 | | SARS-CoV-2 Omicron HV.1 RBD in complex with human ACE2 (local refinement from the spike protein) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-28 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XN5
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron EG.5.1 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNF
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XNK
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron HV.1 spike protein(6P) in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Shi, K.Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2023-12-30 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-27 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8XUS
 
 | | JN.1 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUT
 
 | | XBB.1.5 Spike Trimer in complex with heparan sulfate | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Yue, C, Liu, P, Mao, X. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XUU
 
 | | BA.2.86-T356K Spike Trimer in complex with heparan sulfate (Local refinement) | | Descriptor: | 2-O-sulfo-beta-L-altropyranuronic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Yue, C, Liu, P. | | Deposit date: | 2024-01-14 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Spike N354 glycosylation augments SARS-CoV-2 fitness for human adaptation through structural plasticity.
Natl Sci Rev, 11, 2024
|
|
8XYZ
 
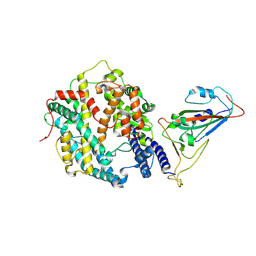 | | The structure of fox ACE2 and PT RBD complex | | Descriptor: | Angiotensin-converting enzyme, Signal peptide, Spike protein S1, ... | | Authors: | sun, J.Q. | | Deposit date: | 2024-01-20 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | The binding and structural basis of fox ACE2 to RBDs from different sarbecoviruses.
Virol Sin, 39, 2024
|
|
8Y16
 
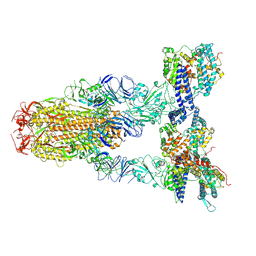 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y5J
 
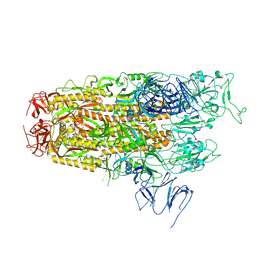 | | Cryo-EM structure of SARS-CoV-2 Omicron JN.1 spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8Y6A
 
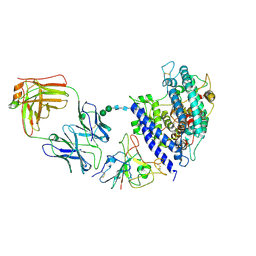 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2.86 RBD in complex with human ACE2 and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | Deposit date: | 2024-02-02 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Spike structures, receptor binding, and immune escape of recently circulating SARS-CoV-2 Omicron BA.2.86, JN.1, EG.5, EG.5.1, and HV.1 sub-variants.
Structure, 32, 2024
|
|
8YRH
 
 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | Authors: | Wang, Q.S, Li, Q.H. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
8VAO
 
 | |
8WZI
 
 | | One RBD up state of Spike glycoprotein, SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yadav, S, Vinothkumar, K.R. | | Deposit date: | 2023-11-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Factors affecting macromolecule orientations in thin films formed in cryo-EM.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8XG2
 
 | | The structure of HLA-A/Pep14 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-14 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XK2
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | Spike protein S1, VHH60 nanobody | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-22 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
8XKC
 
 | | The structure of HLA-A/Pep16 | | Descriptor: | Beta-2-microglobulin, HLA class I heavy chain, Spike protein S1 | | Authors: | Zhang, J.N, Yue, C, Liu, J. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKE
 
 | | The structure of HLA-A/14-3-D | | Descriptor: | Beta-2-microglobulin, GLU-VAL-ASP-ASN-ALA-THR-ARG-PHE-ALA-SER-VAL-TYR, HLA class I heavy chain | | Authors: | Zhang, J.N, Yue, C, Liu, J, Sun, Z.Y. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Uncommon P1 Anchor-featured Viral T Cell Epitope Preference within HLA-A*2601 and HLA-A*0101 Individuals.
Immunohorizons, 8, 2024
|
|
8XKI
 
 | | A neutralizing nanobody VHH60 against wt SARS-CoV-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y, Guo, H, Ji, X, Yang, H. | | Deposit date: | 2023-12-23 | | Release date: | 2024-07-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A broad neutralizing nanobody against SARS-CoV-2 engineered from an approved drug.
Cell Death Dis, 15, 2024
|
|
9AT4
 
 | | Crystal structure of SARS-CoV-2 3CL protease in complex with a methylbicyclo[2.2.1]heptane 2-pyrrolidone inhibitor | | Descriptor: | (1R,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-{[N-({[(2S)-1-{[(1S,2S,4R)-bicyclo[2.2.1]heptan-2-yl]methyl}-5-oxopyrrolidin-2-yl]methoxy}carbonyl)-L-leucyl]amino}-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Dampalla, C.S, Groutas, W.C. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Guided Design of Potent Coronavirus Inhibitors with a 2-Pyrrolidone Scaffold: Biochemical, Crystallographic, and Virological Studies.
J.Med.Chem., 67, 2024
|
|








