8G73
 
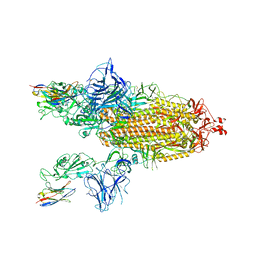 | | SARS-CoV-2 spike/Nb3 complex with 2 RBDs up and 3 Nb3 bound at 2.5 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G74
 
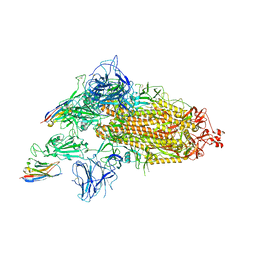 | | SARS-CoV-2 spike/Nb3 complex with 1 RBD up and 2 Nb3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-3, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8G75
 
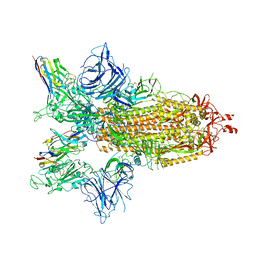 | | SARS-CoV-2 spike/Nb4 complex with 2 RBDs up and 3 Nb4 bound | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanosota-4, ... | | Authors: | Ye, G, Bu, F, Liu, B, Li, F. | | Deposit date: | 2023-02-16 | | Release date: | 2024-01-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Discovery of Nanosota-2, -3, and -4 as super potent and broad-spectrum therapeutic nanobody candidates against COVID-19.
J.Virol., 97, 2023
|
|
8K3K
 
 | |
8U1G
 
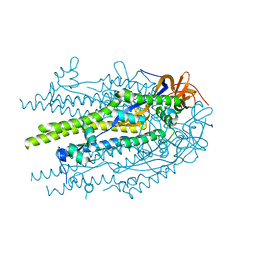 | | Prefusion-stabilized SARS-CoV-2 S2 subunit | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S2 | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2023-08-31 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Prefusion-stabilized SARS-CoV-2 S2-only antigen provides protection against SARS-CoV-2 challenge.
Nat Commun, 15, 2024
|
|
8UH8
 
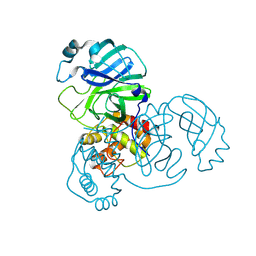 | | Crystal structure of SARS-CoV-2 main protease E166V (Apo structure) | | Descriptor: | ORF1a polyprotein | | Authors: | Bulut, H, Hayashi, H, Kuwata, N, Tsuji, K, Das, D, Tamamura, H, Mitsuya, H. | | Deposit date: | 2023-10-07 | | Release date: | 2024-01-24 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An orally available P1'-5-fluorinated M pro inhibitor blocks SARS-CoV-2 replication without booster and exhibits high genetic barrier.
Pnas Nexus, 4, 2025
|
|
8ENS
 
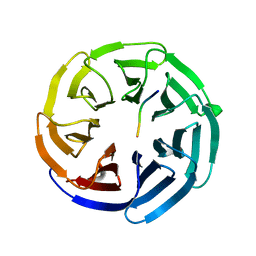 | |
8ENW
 
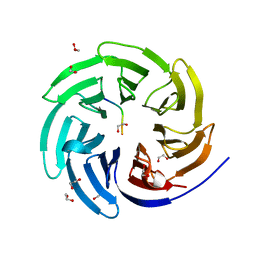 | |
8ENX
 
 | |
8FWO
 
 | | Crystal structure of SARS-CoV-2 papain-like protease | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Bezerra, E.H.S, Soprano, A.S, Tonoli, C.C.C, Prado, P.F.V, da Silva, J.C, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2023-01-23 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 papain-like protease
To Be Published
|
|
8HP9
 
 | |
8HPQ
 
 | |
8HPV
 
 | |
8HQF
 
 | | Crystal structure of SARS-Cov-2 main protease G15S mutant in complex with inhibitor YH-53 | | Descriptor: | N-[(2S)-1-[[(2S)-1-(1,3-benzothiazol-2-yl)-1-oxidanylidene-3-[(3S)-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, Non-structural protein 7 | | Authors: | Zeng, P, Zhang, J, Li, J. | | Deposit date: | 2022-12-13 | | Release date: | 2024-01-31 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Crystal structures of main proteases of SARS-CoV-2 variants bound to a benzothiazole-based inhibitor.
Acta Biochim.Biophys.Sin., 55, 2023
|
|
8I4H
 
 | | Omicron spike variant BA.1 with Bn03 | | Descriptor: | Bn03, Spike glycoprotein | | Authors: | Hao, A.H, Zhang, X, Chen, Z.G, Sun, L. | | Deposit date: | 2023-01-19 | | Release date: | 2024-01-31 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Defining a highly conserved cryptic epitope for antibody recognition of SARS-CoV-2 variants.
Signal Transduct Target Ther, 8, 2023
|
|
8KA8
 
 | | Cryo-EM structure of SARS-CoV-2 Delta RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-02 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8KC2
 
 | | Cryo-EM structure of SARS-CoV-2 BA.3 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, ... | | Authors: | Niu, S, Zhao, Z.N, Chai, Y, Gao, G.F. | | Deposit date: | 2023-08-05 | | Release date: | 2024-01-31 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis and analysis of hamster ACE2 binding to different SARS-CoV-2 spike RBDs.
J.Virol., 98, 2024
|
|
8QPR
 
 | | SARS-CoV-2 S protein bound to human neutralising antibody UZGENT_G5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, IgG heavy chain - FAB, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8QQ0
 
 | | SARS-CoV-2 S protein bound to neutralising antibody UZGENT_A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Remaut, H, Reiter, D, Vandenkerckhove, L, Acar, D.D, Witkowski, W, Gerlo, S. | | Deposit date: | 2023-10-03 | | Release date: | 2024-01-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Integrating artificial intelligence-based epitope prediction in a SARS-CoV-2 antibody discovery pipeline: caution is warranted.
Ebiomedicine, 100, 2024
|
|
8C9L
 
 | | Crystal structure of SARS-CoV-2 Mpro-S144A mutant, free enzyme | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | El Kilani, H, Ibrahim, M, Hilgenfeld, R. | | Deposit date: | 2023-01-23 | | Release date: | 2024-02-07 | | Last modified: | 2025-09-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 13b-K and Nirmatrelvir Resistance Mutations of SARS-CoV-2 Main Protease: Structural, Biochemical, and Biophysical Characterization of Free Enzymes and Inhibitor Complexes
Crystals, 15, 2025
|
|
8CBD
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-1 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-1 heavy chain, BA.4/5-1 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBE
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.4/5-2 and Beta-49 Fabs | | Descriptor: | BA.4/5-2 heavy chain, BA.4/5-2 light chain, Beta-49 heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8CBF
 
 | | SARS-CoV-2 Delta-RBD complexed with Omi-42 and Beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, CHLORIDE ION, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8DT9
 
 | | Crystal Structure of SARS CoV-2 Mpro mutant L141R with Pfizer Intravenous Inhibitor PF-00835231 | | Descriptor: | 3C-like proteinase nsp5, DIMETHYL SULFOXIDE, N-[(2S)-1-({(2S,3S)-3,4-dihydroxy-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide, ... | | Authors: | Shaqra, A.M, Schiffer, C.A. | | Deposit date: | 2022-07-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Contributions of Hyperactive Mutations in M pro from SARS-CoV-2 to Drug Resistance.
Acs Infect Dis., 10, 2024
|
|
8E4W
 
 | |








