9IUQ
 
 | | KP.2 RBD in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Feng, L.L. | | 登録日 | 2024-07-22 | | 公開日 | 2025-01-15 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Cell Discov, 10, 2024
|
|
9IUU
 
 | | JN.1 RBD with Q493E in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, CHLORIDE ION, ... | | 著者 | Feng, L.L. | | 登録日 | 2024-07-22 | | 公開日 | 2025-01-15 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | Structural and molecular basis of the epistasis effect in enhanced affinity between SARS-CoV-2 KP.3 and ACE2.
Cell Discov, 10, 2024
|
|
9JJ7
 
 | |
8XAL
 
 | | Cryo-EM structure of SARS-CoV-2 S-BQ.1 in complex with ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein (Fragment), ... | | 著者 | Hsu, H.F, Wu, M.H, Chang, Y.C, Hsu, S.T.D. | | 登録日 | 2023-12-04 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Functional and structural investigation of a broadly neutralizing SARS-CoV-2 antibody.
JCI Insight, 9, 2024
|
|
8XWR
 
 | |
8XWT
 
 | |
8XY9
 
 | | Crystal structure of SARS-CoV-2 BF.7 RBD and human ACE2 complex | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Lan, J, Wang, C.H. | | 登録日 | 2024-01-19 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-04-16 | | 実験手法 | X-RAY DIFFRACTION (3.64 Å) | | 主引用文献 | Receptor binding mechanism and immune evasion capacity of SARS-CoV-2 BQ.1.1 lineage.
Virology, 600, 2024
|
|
8XYE
 
 | | Crystal structure of SARS-CoV-2 BA.4 RBD and human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | 著者 | Lan, J, Wang, C.H. | | 登録日 | 2024-01-19 | | 公開日 | 2025-01-22 | | 最終更新日 | 2025-04-16 | | 実験手法 | X-RAY DIFFRACTION (3.32 Å) | | 主引用文献 | Receptor binding mechanism and immune evasion capacity of SARS-CoV-2 BQ.1.1 lineage.
Virology, 600, 2024
|
|
8XYG
 
 | |
8XYO
 
 | | Cryo-EM structure of CX1 receptor binding domain in complex with human ACE2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein, ... | | 著者 | Xu, Z.P, Li, L.J, Gu, Y.H, Qi, J.X, Gao, G.F. | | 登録日 | 2024-01-20 | | 公開日 | 2025-01-22 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Structural resemblances of bat-origin coronaviruses CX1 and BANAL-20-52 spikes provide further evidence on bat origination of SARS-CoV-2
To Be Published
|
|
8YZC
 
 | | Structure of BA.2.86 spike protein in complex with ACE2. | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | 著者 | Wang, Y.J, Zang, X, Sun, L. | | 登録日 | 2024-04-06 | | 公開日 | 2025-01-22 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
8YZE
 
 | | The JN.1 spike protein (S) in complex with ACE2. | | 分子名称: | Angiotensin-converting enzyme 2, Spike glycoprotein,Fibritin,Expression Tag | | 著者 | Wang, Y.J, Zhang, X, Sun, L. | | 登録日 | 2024-04-06 | | 公開日 | 2025-01-22 | | 実験手法 | ELECTRON MICROSCOPY (3.06 Å) | | 主引用文献 | Lineage-specific pathogenicity, immune evasion, and virological features of SARS-CoV-2 BA.2.86/JN.1 and EG.5.1/HK.3.
Nat Commun, 15, 2024
|
|
9EXA
 
 | |
9JS4
 
 | |
8YZR
 
 | |
8YZW
 
 | |
8YZZ
 
 | | The structure of HLA-A*2402 complex with peptide from SARS-CoV-2 S448-456 NYNYLYRLF(Prototype) | | 分子名称: | Beta-2-microglobulin, MHC class I antigen, Spike protein S1 | | 著者 | Shang, B.L, Zhang, J.N, Tian, J.M, Liu, J. | | 登録日 | 2024-04-08 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
8Z05
 
 | | The structure of HLA-A*0201 complex with peptide from SARS-CoV-2 N222-230 LLLDRLNKL(BA.2.86/JN.1) | | 分子名称: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | 著者 | Zhang, J.N, Tian, J.M, Liu, J. | | 登録日 | 2024-04-09 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | T cell immune evasion by SARS-CoV-2 JN.1 escapees targeting two cytotoxic T cell epitope hotspots.
Nat.Immunol., 26, 2025
|
|
8Z06
 
 | |
8Z07
 
 | |
8Z08
 
 | |
8Z6R
 
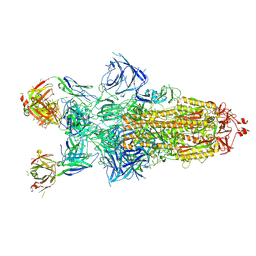 | | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1. | | 分子名称: | CYFN1006-1 heavy chain, CYFN1006-1 light chain, Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Spike glycoprotein,Fibritin,Expression Tag | | 著者 | Wang, Y.J, Sun, L. | | 登録日 | 2024-04-19 | | 公開日 | 2025-01-29 | | 実験手法 | ELECTRON MICROSCOPY (2.87 Å) | | 主引用文献 | Structure of XBB.1.16 S trimer with 3 down-RBDs complex with antibody CYFN1006-1.
To Be Published
|
|
8Z6X
 
 | |
9ARU
 
 | | COVA2-15 fragment antigen binding in complex with SARS-CoV-2 6P-mut7 S protein | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COVA2-15 heavy chain variable region, ... | | 著者 | Ozorowski, G, Turner, H.L, Ward, A.B. | | 登録日 | 2024-02-23 | | 公開日 | 2025-01-29 | | 最終更新日 | 2025-05-28 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Plant-produced SARS-CoV-2 antibody engineered towards enhanced potency and in vivo efficacy.
Plant Biotechnol J, 23, 2025
|
|
9G9I
 
 | |








