8DMA
 
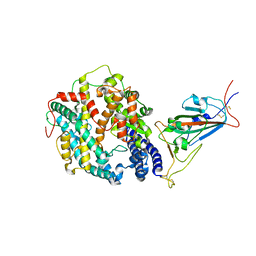 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.1 spike protein in complex with mouse ACE2 (focused refinement of RBD and ACE2) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Spike glycoprotein | | 著者 | Zhu, X, Saville, J.W, Mannar, D, Berezuk, A.M, Cholak, S, Tuttle, K.S, Vahdatihassani, F, Subramaniam, S. | | 登録日 | 2022-07-08 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-11-20 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein.
Cell Rep, 42, 2023
|
|
8DQU
 
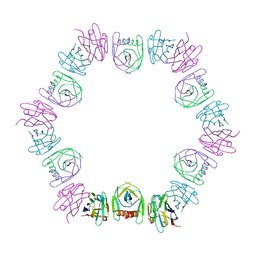 | | Nanobody bound SARS-CoV-2 Nsp9 | | 分子名称: | Nanobody, Non-structural protein 9 | | 著者 | Littler, D.R, Gully, B.S, Rossjohn, J. | | 登録日 | 2022-07-20 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-11-06 | | 実験手法 | X-RAY DIFFRACTION (2.45003185971 Å) | | 主引用文献 | Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9.
Plos One, 18, 2023
|
|
8EQJ
 
 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like membrane environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-07 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQT
 
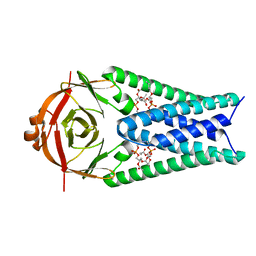 | | Structure of SARS-CoV-2 Orf3a in plasma membrane-like environment, MSP1D1 nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-09 | | 公開日 | 2023-02-08 | | 最終更新日 | 2024-06-19 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
8EQU
 
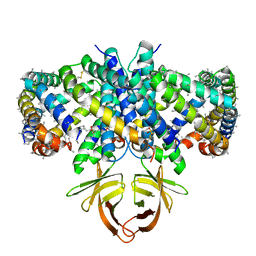 | | Structure of SARS-CoV-2 Orf3a in late endosome/lysosome-like environment, Saposin A nanodisc | | 分子名称: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ORF3a protein, Saposin A, ... | | 著者 | Miller, A.N, Houlihan, P.R, Matamala, E, Cabezas-Bratesco, D, Lee, G.Y, Cristofori-Armstrong, B, Dilan, T.L, Sanchez-Martinez, S, Matthies, D, Yan, R, Yu, Z, Ren, D, Brauchi, S.E, Clapham, D.E. | | 登録日 | 2022-10-09 | | 公開日 | 2023-02-08 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | The SARS-CoV-2 accessory protein Orf3a is not an ion channel, but does interact with trafficking proteins.
Elife, 12, 2023
|
|
7WWI
 
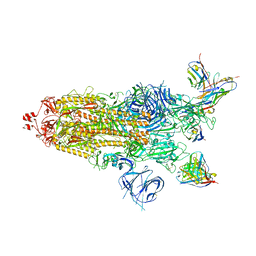 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 1 conformation
To Be Published
|
|
7WWJ
 
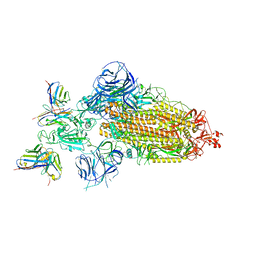 | | SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab in the class 2 conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 55A8 heavy chain, ... | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | EM structure of SARS-CoV-2 Omicron variant spike glycoprotein and 55A8
To Be Published
|
|
7WWK
 
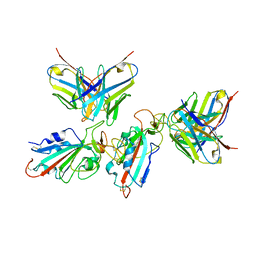 | | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab | | 分子名称: | 55A8 light chain, Spike glycoprotein | | 著者 | Guo, H, Gao, Y, Lu, Y, Yang, H, Ji, X. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Local refinement of the SARS-CoV-2 BA.1 Spike trimer in complex with 55A8 Fab
To Be Published
|
|
7Y7J
 
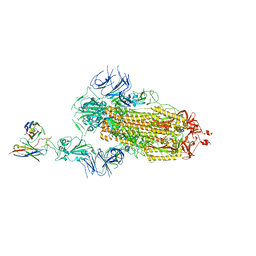 | | SARS-CoV-2 S trimer in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
7Y7K
 
 | | SARS-CoV-2 RBD in complex with 1F Fab | | 分子名称: | 1F VH, 1F VL, Spike protein S1 | | 著者 | Zhao, S, Liu, F, Yang, X, Zhong, G. | | 登録日 | 2022-06-22 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | A core epitope targeting antibody of SARS-CoV-2.
Protein Cell, 14, 2023
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | 分子名称: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | 著者 | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | 登録日 | 2022-09-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-11-13 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
8C8P
 
 | |
8G62
 
 | | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004 | | 分子名称: | 3-methoxy-5-(1-methylpiperidin-4-yl)-N-[4-(pyrrolidine-1-sulfonyl)phenyl]benzamide, ACETATE ION, CHLORIDE ION, ... | | 著者 | Osipiuk, J, Tesar, C, Endres, M, Jedrzejczak, R, Luci, D, Kales, S, Simeonov, A, Rai, G, Drayman, N, Tay, S, Oakes, S, Rosner, M, Chen, B, Dulin, N, Solway, J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | 登録日 | 2023-02-14 | | 公開日 | 2023-02-22 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Papain-Like Protease of SARS CoV-2 in complex with remodilin NCGC 390004
To Be Published
|
|
7U0Q
 
 | |
7U0X
 
 | |
7WP6
 
 | | Cryo-EM structure of SARS-CoV-2 recombinant spike protein STFK in complex with three neutralizing antibodies | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 36H6 heavy chain, ... | | 著者 | Zheng, Q, Sun, H, Yuan, Q, Li, S, Xia, N. | | 登録日 | 2022-01-23 | | 公開日 | 2023-03-01 | | 最終更新日 | 2025-06-18 | | 実験手法 | ELECTRON MICROSCOPY (3.81 Å) | | 主引用文献 | Lineage-mosaic and mutation-patched spike proteins for broad-spectrum COVID-19 vaccine.
Cell Host Microbe, 30, 2022
|
|
7X2H
 
 | |
7XCZ
 
 | |
7XDA
 
 | |
7XDB
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron Spike protein in complex with BA7208 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7208 fab, ... | | 著者 | Liu, Z, Liu, S, Gao, Y.Z. | | 登録日 | 2022-03-26 | | 公開日 | 2023-03-01 | | 最終更新日 | 2025-06-25 | | 実験手法 | ELECTRON MICROSCOPY (2.62 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XDK
 
 | | Cryo-EM structure of SARS-CoV-2 Delta Spike protein in complex with BA7054 and BA7125 fab | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA7054 fab, ... | | 著者 | Liu, Z, Lui, S, Gao, Y. | | 登録日 | 2022-03-27 | | 公開日 | 2023-03-01 | | 最終更新日 | 2025-07-02 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Biparatopic antibody BA7208/7125 effectively neutralizes SARS-CoV-2 variants including Omicron BA.1-BA.5.
Cell Discov, 9, 2023
|
|
7XQ6
 
 | | The complex structure of mutant Mpro with inhibitor | | 分子名称: | 3C-like proteinase nsp5, CHLORIDE ION | | 著者 | Sahoo, P, Lenka, D.R, Kumar, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7XQ7
 
 | | The complex structure of WT-Mpro | | 分子名称: | 3C-like proteinase nsp5, SODIUM ION | | 著者 | Sahoo, P, Lenka, D.R, Kumar, A. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Detailed Insights into the Inhibitory Mechanism of New Ebselen Derivatives against Main Protease (M pro ) of Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2).
Acs Pharmacol Transl Sci, 6, 2023
|
|
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-06 | | 公開日 | 2023-03-01 | | 最終更新日 | 2023-05-24 | | 実験手法 | ELECTRON MICROSCOPY (3.29 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | 著者 | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | 登録日 | 2022-05-08 | | 公開日 | 2023-03-01 | | 最終更新日 | 2024-11-13 | | 実験手法 | ELECTRON MICROSCOPY (2.63 Å) | | 主引用文献 | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|








