登録情報 データベース : PDB / ID : 7jhgタイトル Cryo-EM structure of ATP-bound fully inactive AMPK in complex with Dorsomorphin (Compound C) and Fab-nanobody (5'-AMP-activated protein kinase subunit ...) x 2 5'-AMP-activated protein kinase catalytic subunit alpha-1 Fab heavy chain Fab light chain Maltodextrin-binding protein Nanobody キーワード / / / / / 機能・相同性 分子機能 ドメイン・相同性 構成要素
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / 生物種 Homo sapiens (ヒト)Escherichia coli K-12 (大腸菌)synthetic construct (人工物) 手法 / / / 解像度 : 3.47 Å データ登録者 Yan, Y. / Murkherjee, S. / Zhou, X.E. / Xu, T.H. / Xu, H.E. / Kossiakoff, A.A. / Melcher, K. 資金援助 組織 認可番号 国 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R01 GM117372 National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) R01 GM129436
ジャーナル : Science / 年 : 2021タイトル : Structure of an AMPK complex in an inactive, ATP-bound state.著者: Yan Yan / Somnath Mukherjee / Kaleeckal G Harikumar / Timothy S Strutzenberg / X Edward Zhou / Kelly Suino-Powell / Ting-Hai Xu / Ryan D Sheldon / Jared Lamp / Joseph S Brunzelle / Katarzyna ... 著者 : Yan Yan / Somnath Mukherjee / Kaleeckal G Harikumar / Timothy S Strutzenberg / X Edward Zhou / Kelly Suino-Powell / Ting-Hai Xu / Ryan D Sheldon / Jared Lamp / Joseph S Brunzelle / Katarzyna Radziwon / Abigail Ellis / Scott J Novick / Irving E Vega / Russell G Jones / Laurence J Miller / H Eric Xu / Patrick R Griffin / Anthony A Kossiakoff / Karsten Melcher / 要旨 : Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in ... Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility. 履歴 登録 2020年7月20日 登録サイト / 処理サイト 改定 1.0 2021年7月21日 Provider / タイプ 改定 1.1 2021年12月15日 Group / カテゴリ / citation_author / database_2Item _citation.country / _citation.journal_abbrev ... _citation.country / _citation.journal_abbrev / _citation.journal_id_ASTM / _citation.journal_id_CSD / _citation.journal_id_ISSN / _citation.journal_volume / _citation.page_first / _citation.page_last / _citation.pdbx_database_id_DOI / _citation.pdbx_database_id_PubMed / _citation.title / _citation.year / _citation_author.identifier_ORCID / _citation_author.name / _database_2.pdbx_DOI / _database_2.pdbx_database_accession 改定 1.2 2024年11月6日 Group / Refinement description / Structure summaryカテゴリ chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / em_3d_fitting_list / em_admin / pdbx_entry_details / pdbx_initial_refinement_model / pdbx_modification_feature Item _em_3d_fitting_list.accession_code / _em_3d_fitting_list.initial_refinement_model_id ... _em_3d_fitting_list.accession_code / _em_3d_fitting_list.initial_refinement_model_id / _em_3d_fitting_list.source_name / _em_3d_fitting_list.type / _em_admin.last_update / _pdbx_entry_details.has_protein_modification
すべて表示 表示を減らす
 データを開く
データを開く 基本情報
基本情報 要素
要素 キーワード
キーワード 機能・相同性情報
機能・相同性情報 Homo sapiens (ヒト)
Homo sapiens (ヒト)
 データ登録者
データ登録者 米国, 2件
米国, 2件  引用
引用 ジャーナル: Science / 年: 2021
ジャーナル: Science / 年: 2021

 構造の表示
構造の表示 ムービービューア
ムービービューア Molmil
Molmil Jmol/JSmol
Jmol/JSmol ダウンロードとリンク
ダウンロードとリンク ダウンロード
ダウンロード 7jhg.cif.gz
7jhg.cif.gz PDBx/mmCIF形式
PDBx/mmCIF形式 pdb7jhg.ent.gz
pdb7jhg.ent.gz PDB形式
PDB形式 7jhg.json.gz
7jhg.json.gz PDBx/mmJSON形式
PDBx/mmJSON形式 その他のダウンロード
その他のダウンロード 7jhg_validation.pdf.gz
7jhg_validation.pdf.gz wwPDB検証レポート
wwPDB検証レポート 7jhg_full_validation.pdf.gz
7jhg_full_validation.pdf.gz 7jhg_validation.xml.gz
7jhg_validation.xml.gz 7jhg_validation.cif.gz
7jhg_validation.cif.gz https://data.pdbj.org/pub/pdb/validation_reports/jh/7jhg
https://data.pdbj.org/pub/pdb/validation_reports/jh/7jhg ftp://data.pdbj.org/pub/pdb/validation_reports/jh/7jhg
ftp://data.pdbj.org/pub/pdb/validation_reports/jh/7jhg リンク
リンク 集合体
集合体
 要素
要素 Homo sapiens (ヒト) / 遺伝子: PRKAA1, AMPK1 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: PRKAA1, AMPK1 / 発現宿主: 


 Homo sapiens (ヒト) / 遺伝子: PRKAB2 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: PRKAB2 / 発現宿主: 
 Homo sapiens (ヒト) / 遺伝子: PRKAG1 / 発現宿主:
Homo sapiens (ヒト) / 遺伝子: PRKAG1 / 発現宿主: 



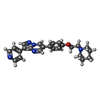






 試料調製
試料調製 電子顕微鏡撮影
電子顕微鏡撮影
 FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM
FIELD EMISSION GUN / 加速電圧: 300 kV / 照射モード: FLOOD BEAM 解析
解析 ムービー
ムービー コントローラー
コントローラー















 PDBj
PDBj





























